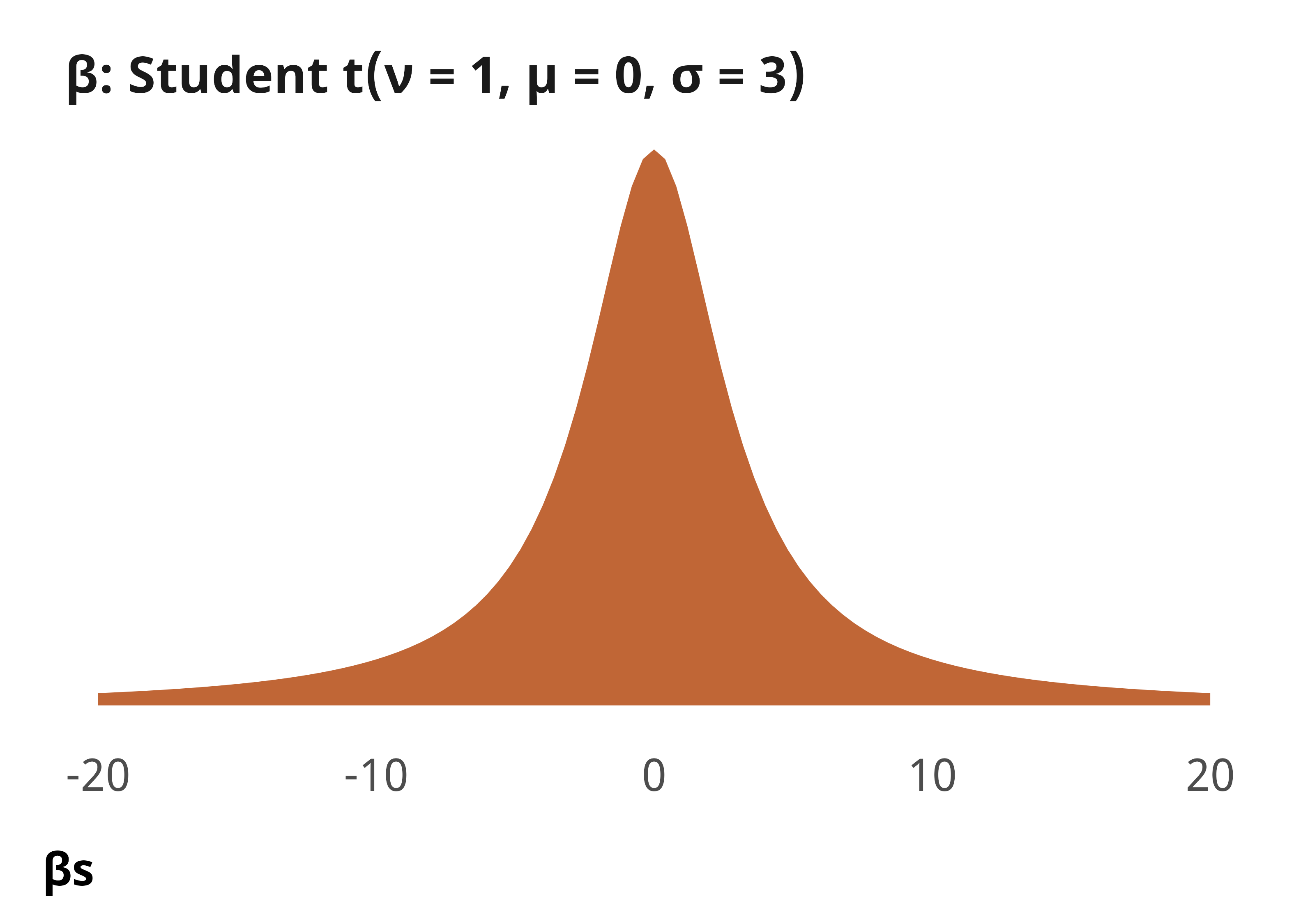
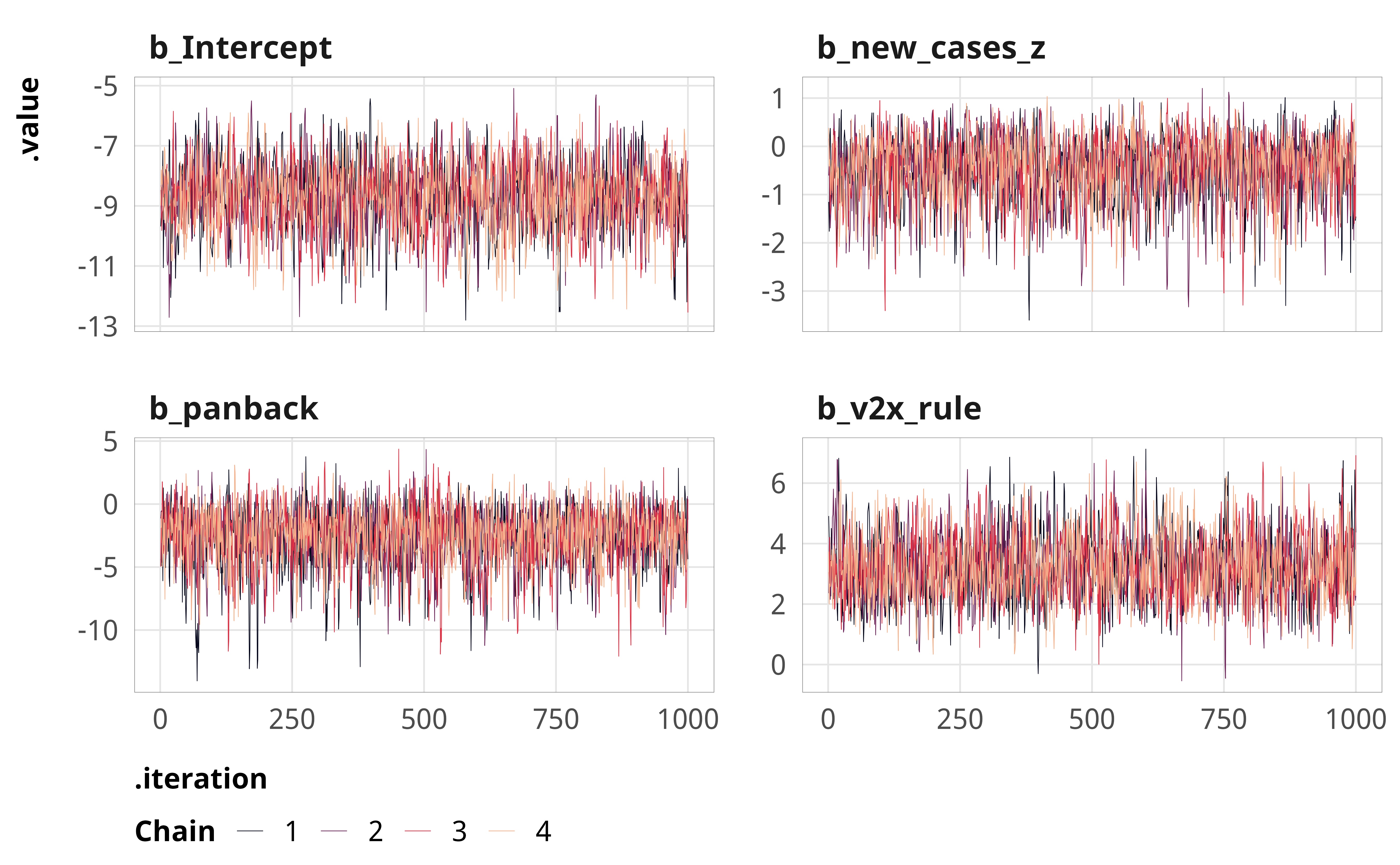
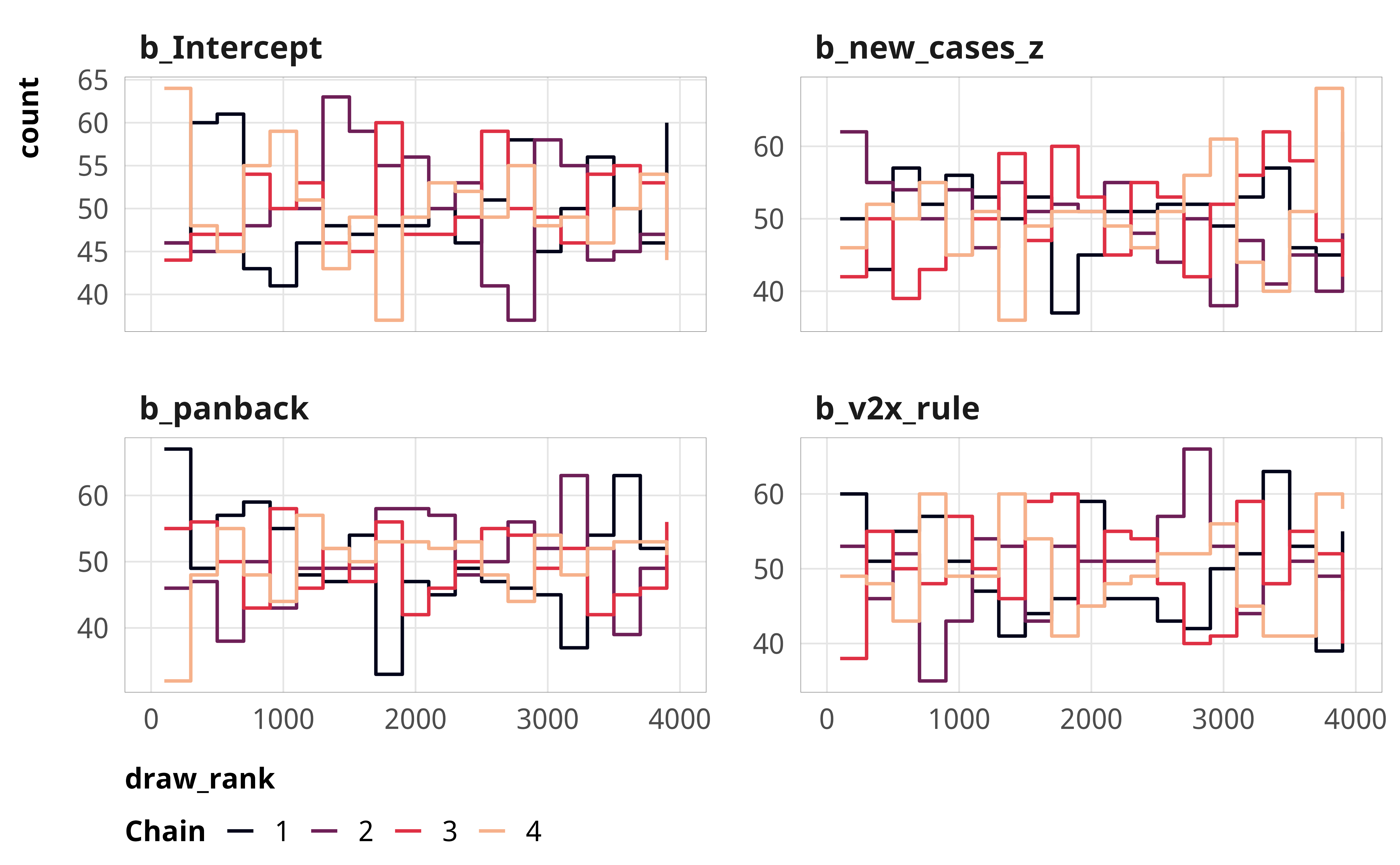
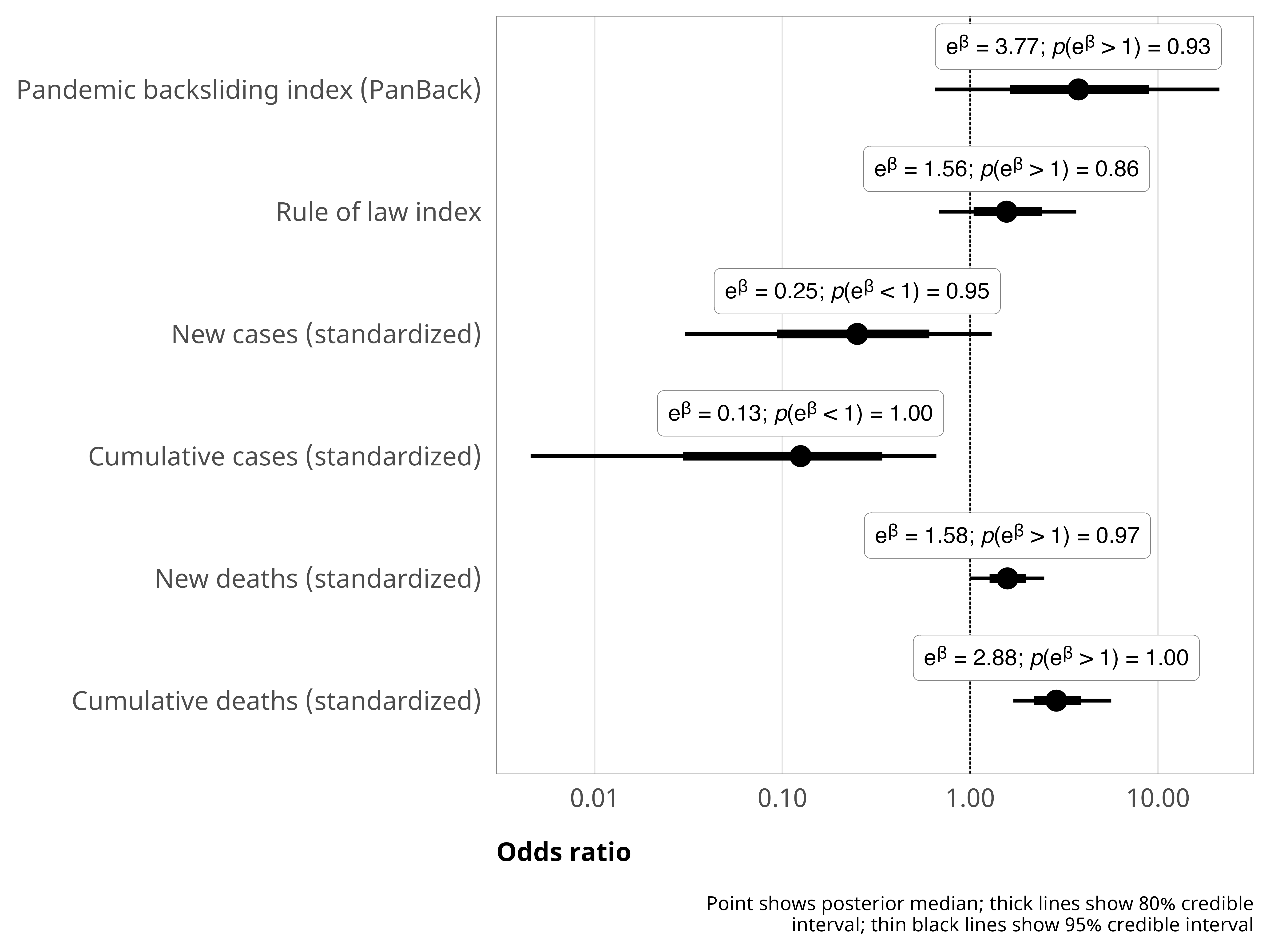
--- title: "Explaining derogations" format: html: code-fold: true --- ```{r} #| label: setup #| include: false :: opts_chunk$ set (fig.width = 6 , fig.height = (6 * 0.618 ), out.width = "80%" ,fig.align = "center" , fig.retina = 3 ,collapse = TRUE options (digits = 3 , width = 120 , dplyr.summarise.inform = FALSE )``` ```{r} #| label: libraries-data #| warning: false #| message: false library (tidyverse)library (tidybayes)library (modelsummary)library (scales)library (glue)library (ggtext)library (tinytable)library (targets)tar_config_set (store = here:: here ("_targets" ),script = here:: here ("_targets.R" )tar_load (c (m_derogations, m_tbl_derogations, action_state_type))invisible (list2env (tar_read (graphic_functions), .GlobalEnv))invisible (list2env (tar_read (diagnostic_functions), .GlobalEnv))invisible (list2env (tar_read (helper_functions), .GlobalEnv))invisible (list2env (tar_read (modelsummary_functions), .GlobalEnv))``` # Model details ## Formal model specification \\ \\ [ 0.75em ] \\ \\ \\ \\ \\ [ 0.75em ] \\ ## Priors ```{r} #| label: figure-prior #| fig-cap: "Density plot of prior distribution for model parameters" #| fig-width: 3.5 #| fig-height: 2.5 #| out-width: "50%" ggplot () + stat_function (geom = "area" ,fun = ~ extraDistr:: dlst (., df = 1 , mu = 0 , sigma = 3 ),fill = clrs[2 ]+ xlim (c (- 20 , 20 )) + labs (x = "βs" ) + facet_wrap (vars ("β: Student t(ν = 1, µ = 0, σ = 3)" )) + theme_pandem (prior = TRUE )``` ## Simplified R code ```r brm (bf (outcome ~ panback + + cumulative_cases_z + + cumulative_deaths_z + + year_week_num),family = bernoulli (),prior = c (prior (student_t (1 , 0 , 3 ), class = Intercept),prior (student_t (1 , 0 , 3 ), class = b)),``` ## Model evaluation ```{r} <- c ("b_Intercept" , "b_panback" , "b_new_cases_z" , "b_v2x_rule" )``` ### Derogation filed ```{r} plot_trace (m_derogations$ m_derogations_panback, params_to_show)``` ```{r} plot_trank (m_derogations$ m_derogations_panback, params_to_show)``` ```{r} plot_pp (m_derogations$ m_derogations_panback)``` ### Other treaty action ```{r} plot_trace (m_derogations$ m_other_panback, params_to_show)``` ```{r} plot_trank (m_derogations$ m_other_panback, params_to_show)``` ```{r} plot_pp (m_derogations$ m_other_panback)``` # Results ```{r} #| label: calc-derog-coefs <- tribble (~ coef, ~ coef_nice,"b_panback" , "Pandemic backsliding index (PanBack)" ,"b_v2x_rule" , "Rule of law index" ,"b_new_cases_z" , "New cases (standardized)" ,"b_cumulative_cases_z" , "Cumulative cases (standardized)" ,"b_new_deaths_z" , "New deaths (standardized)" ,"b_cumulative_deaths_z" , "Cumulative deaths (standardized)" |> mutate (coef_nice = fct_inorder (coef_nice))<- m_derogations$ m_derogations_panback |> gather_draws (` ^b_.* ` , regex = TRUE ) |> filter (.variable %in% coef_lookup$ coef) |> left_join (coef_lookup, by = join_by (.variable == coef))<- m_derog_draws |> mutate (.value_exp = exp (.value)) |> group_by (.variable, coef_nice) |> reframe (post_medians = median_hdci (.value_exp, .width = 0.95 ),p_gt_0 = sum (.value_exp > 1 ) / n ()|> unnest (post_medians) |> mutate (y_nice = fmt_coef (y),y_nice_html = fmt_coef (y, html = TRUE )|> mutate (p_lt_0 = 1 - p_gt_0,p_gt = fmt_p_inline (p_gt_0, "gt" ),p_lt = fmt_p_inline (p_lt_0, "lt" ),p_gt_html = fmt_p_inline (p_gt_0, "gt" , html = TRUE ),p_lt_html = fmt_p_inline (p_lt_0, "lt" , html = TRUE )|> mutate (p_d = if_else (y > 1 , p_gt, p_lt),p_d_html = if_else (y > 1 , p_gt_html, p_lt_html),plot_label = glue ("{y_nice_html}; {p_d_html}" )|> mutate (or_pct = label_percent (accuracy = 1 )(abs (1 - y)))``` ## Coefficient plot ```{r} #| label: figure-derog-coefs #| fig-width: 6 #| fig-height: 4.5 #| fig-cap: "Odds ratios for coefficients from logistic regression model predicting the probability of derogation from the ICCPR" #| out-width: 100% |> mutate (.value = exp (.value)) |> ggplot (aes (x = .value, y = fct_rev (coef_nice))) + stat_pointinterval () + geom_vline (xintercept = 1 , linewidth = 0.25 , linetype = "21" ) + geom_richtext (data = derog_coefs, aes (x = y, label = plot_label),size = 2.7 , nudge_y = 0.35 , label.size = 0.1 , label.colour = "grey50" + scale_x_log10 () + labs (x = "Odds ratio" , y = NULL ,caption = str_wrap (glue ("Point shows posterior median;" , " thick lines show 80% credible interval;" ,"thin black lines show 95% credible interval" ,.sep = " " width = 60 + theme_pandem () + theme (panel.grid.major.y = element_blank ())``` ## Complete table of results ```{r} #| label: table-results-full-derogations #| tbl-cap: "Complete results from models showing predictors of derogations (H~1~)" <- paste ("Note: Estimates are median posterior log odds from logistic regression models;" ,"95% credible intervals (highest density posterior interval, or HDPI) in brackets." |> set_names ("Derogation filed" , "Other action" ) |> modelsummary (estimate = "{estimate}" ,statistic = "[{conf.low}, {conf.high}]" ,coef_map = coef_map,gof_map = gof_map,output = "tinytable" ,fmt = fmt_significant (2 ),notes = c (notes),width = c (0.4 , rep (0.3 , 2 ))|> group_tt (j = list ("ICCPR action" = 2 : 3 )) |> style_tt (j = 2 : 3 , align = "c" ) |> style_tt (i = seq (1 , 15 , 2 ), j = 1 , rowspan = 2 , alignv = "t" ) |> style_tt (bootstrap_class = "table table-sm" ```