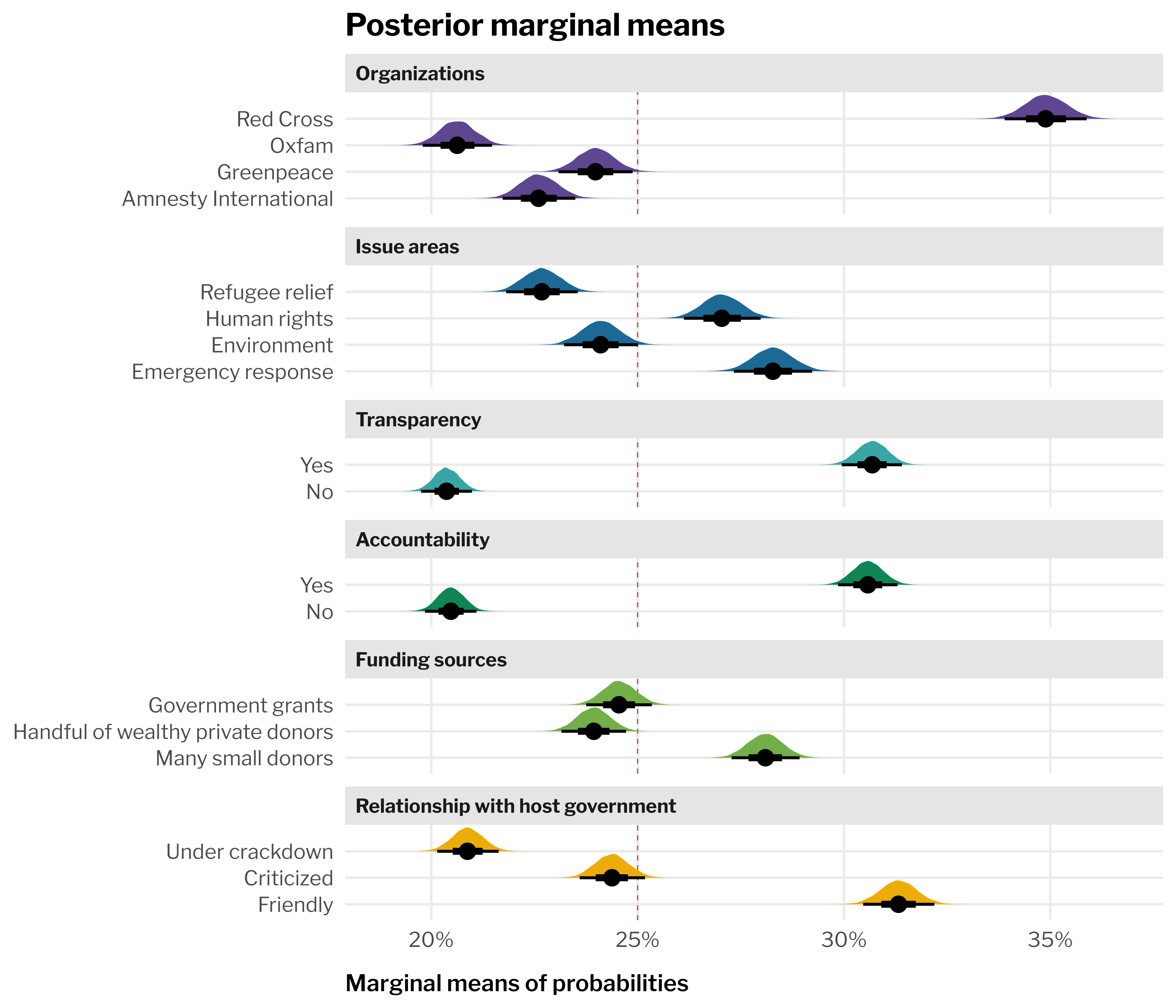
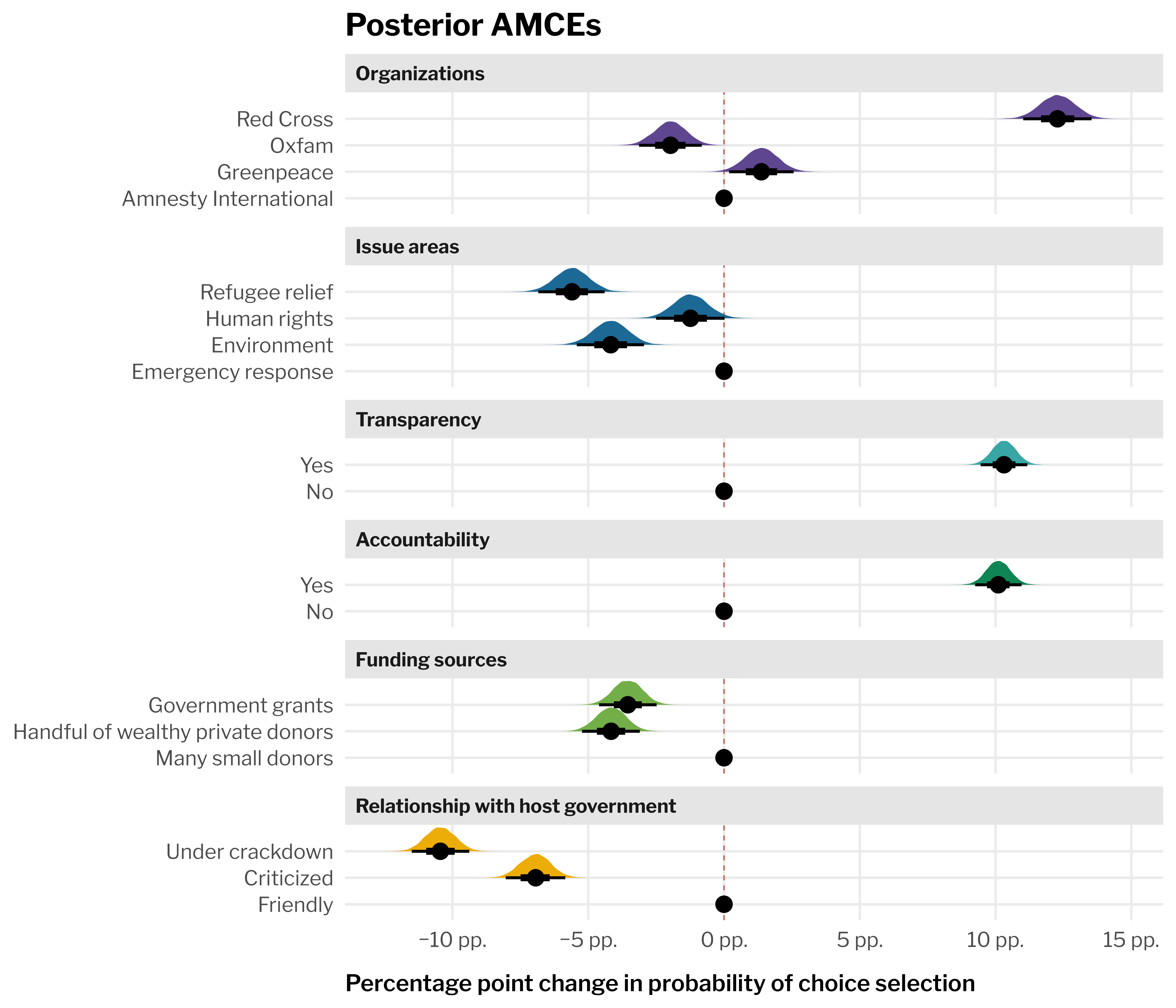
--- title: "All MMs and AMCEs" format: html: code-fold: true --- ```{r setup, include=FALSE} :: opts_chunk$ set (fig.align = "center" , fig.retina = 3 ,fig.width = 6 , fig.height = (6 * 0.618 ),out.width = "80%" , collapse = TRUE ,dev = "ragg_png" options (digits = 3 , width = 120 ,dplyr.summarise.inform = FALSE ,knitr.kable.NA = "" ``` ```{r libraries-data, warning=FALSE, message=FALSE} library (tidyverse)library (targets)library (tidybayes)library (patchwork)library (ggforce)library (scales)library (glue)library (gt)library (gtExtras)library (here)# Targets stuff tar_config_set (store = here ('_targets' ),script = here ('_targets.R' ))tar_load (c (grid_treatment_only, level_lookup, feature_lookup))<- tar_read (preds_conditional_treatment_only)invisible (list2env (tar_read (graphic_functions), .GlobalEnv))invisible (list2env (tar_read (table_functions), .GlobalEnv))theme_set (theme_ngo ())``` ```{r calc-mms} <- preds_all %>% group_by (feat_org, .draw) %>% summarize (avg = mean (.epred))<- preds_all %>% group_by (feat_issue, .draw) %>% summarize (avg = mean (.epred))<- preds_all %>% group_by (feat_transp, .draw) %>% summarize (avg = mean (.epred))<- preds_all %>% group_by (feat_acc, .draw) %>% summarize (avg = mean (.epred))<- preds_all %>% group_by (feat_funding, .draw) %>% summarize (avg = mean (.epred))<- preds_all %>% group_by (feat_govt, .draw) %>% summarize (avg = mean (.epred))``` ```{r calc-amces} <- mms_org %>% group_by (feat_org) %>% compare_levels (variable = avg, by = feat_org, comparison = "control" ) %>% ungroup () %>% separate_wider_delim (delim = " - " , names = c ("feature_level" , "reference_level" )%>% add_row (avg = 0 , feature_level = unique (.$ reference_level))<- mms_issue %>% group_by (feat_issue) %>% compare_levels (variable = avg, by = feat_issue, comparison = "control" ) %>% ungroup () %>% separate_wider_delim (delim = " - " , names = c ("feature_level" , "reference_level" )%>% add_row (avg = 0 , feature_level = unique (.$ reference_level))<- mms_transp %>% group_by (feat_transp) %>% compare_levels (variable = avg, by = feat_transp, comparison = "control" ) %>% ungroup () %>% separate_wider_delim (delim = " - " , names = c ("feature_level" , "reference_level" )%>% add_row (avg = 0 , feature_level = unique (.$ reference_level))<- mms_acc %>% group_by (feat_acc) %>% compare_levels (variable = avg, by = feat_acc, comparison = "control" ) %>% ungroup () %>% separate_wider_delim (delim = " - " , names = c ("feature_level" , "reference_level" )%>% add_row (avg = 0 , feature_level = unique (.$ reference_level))<- mms_funding %>% group_by (feat_funding) %>% compare_levels (variable = avg, by = feat_funding, comparison = "control" ) %>% ungroup () %>% separate_wider_delim (delim = " - " , names = c ("feature_level" , "reference_level" )%>% add_row (avg = 0 , feature_level = unique (.$ reference_level))<- mms_govt %>% group_by (feat_govt) %>% compare_levels (variable = avg, by = feat_govt, comparison = "control" ) %>% ungroup () %>% separate_wider_delim (delim = " - " , names = c ("feature_level" , "reference_level" )%>% add_row (avg = 0 , feature_level = unique (.$ reference_level))<- bind_rows (lst (amces_org, amces_issue, amces_transp, amces_acc, amces_funding, amces_govt),.id = "amce_var" %>% left_join (select (feature_lookup, amce_var, feature_nice), by = join_by (amce_var)) %>% left_join (select (level_lookup, contains ("_level" )), by = join_by (feature_level)``` ```{r plot-mms-all, fig.width=7, fig.height=6, out.width="100%"} <- bind_rows (lst (mms_org = rename (mms_org, feature_level = feat_org), mms_issue = rename (mms_issue, feature_level = feat_issue), mms_transp = rename (mms_transp, feature_level = feat_transp), mms_acc = rename (mms_acc, feature_level = feat_acc), mms_funding = rename (mms_funding, feature_level = feat_funding), mms_govt = rename (mms_govt, feature_level = feat_govt).id = "mm_var" %>% left_join (select (feature_lookup, mm_var, feature_nice), by = join_by (mm_var)) %>% left_join (select (level_lookup, contains ("_level" )), by = join_by (feature_level)ggplot (aes (x = avg, y = feature_short_level, fill = feature_nice)+ geom_vline (xintercept = 0.25 , color = clrs$ prism[8 ], linetype = "dashed" , linewidth = 0.25 ) + stat_halfeye (normalize = "groups" ) + guides (fill = "none" ) + facet_col (facets = "feature_nice" , scales = "free_y" , space = "free" ) + scale_x_continuous (labels = label_percent ()) + scale_fill_manual (values = clrs$ prism[1 : 6 ]) + labs (x = "Marginal means of probabilities" ,y = NULL ,title = "Posterior marginal means" ``` ```{r plot-amces-all, fig.width=7, fig.height=6, out.width="100%"} ggplot (aes (x = avg, y = feature_short_level, fill = feature_nice)+ geom_vline (xintercept = 0 , color = clrs$ prism[8 ], linetype = "dashed" , linewidth = 0.25 ) + stat_halfeye (normalize = "groups" ) + # Make the heights of the distributions equal within each facet guides (fill = "none" ) + facet_col (facets = "feature_nice" , scales = "free_y" , space = "free" ) + scale_x_continuous (labels = label_pp) + scale_fill_manual (values = clrs$ prism[1 : 6 ]) + labs (x = "Percentage point change in probability of choice selection" ,y = NULL ,title = "Posterior AMCEs" ```