Get, clean, and merge data
Suparna Chaudhry and Andrew Heiss
2018-10-29
knitr::opts_chunk$set(fig.retina = 2,
tidy.opts = list(width.cutoff = 120), # For code
options(width = 120)) # For output
library(tidyverse)
library(stringr)
library(lubridate)
library(httr)
library(rvest)
library(countrycode)
library(readxl)
library(DT)
library(WDI)
library(xml2)
library(ggstance)
library(Amelia)
library(imputeTS)
library(validate)
library(here)
source(here("lib", "graphics.R"))
source(here("lib", "pandoc.R"))
all.na.means.zero <- TRUE
my.seed <- 1234
set.seed(my.seed)Consistent country names
This is a perpetual nightmare in IR data. Here’s a master lookup table of COW codes, ISO3 codes, ISO2 codes, and Gleditsch Ward codes.
Gleditsch and Ward do not include a bunch of countries, so I create my own codes for relevant countries that appear in other datasets:
- Somaliland: 521
- Palestine (West Bank): 667
- Palestine (Gaza): 668
- Palestine (both): 669
- Hong Kong: 715
Also, following Gleditsch and Ward, I treat Serbia as 340 and Serbia & Montenegro as a continuation of Yugoslavia, or 345.
Also, following V-Dem and Polity, I treat Vietnam as 816.
# Gleditsch Ward codes
#
# Download and cache data
gw.states.url <- "https://web.archive.org/web/20170302085046/http://privatewww.essex.ac.uk/~ksg/data/iisystem.dat"
gs.microstates.url <- "https://web.archive.org/web/20170302072201/http://privatewww.essex.ac.uk/~ksg/data/microstatessystem.dat"
gw.states.path <- here("Data", "data_raw", "Gleditsch Ward")
if (!file.exists(file.path(gw.states.path, basename(gw.states.url))) |
!file.exists(file.path(gw.states.path, basename(gs.microstates.url)))) {
GET(gw.states.url,
write_disk(file.path(gw.states.path, basename(gw.states.url)),
overwrite = TRUE))
GET(gs.microstates.url,
write_disk(file.path(gw.states.path, basename(gs.microstates.url)),
overwrite = TRUE))
}
gw.date <- "%d:%m:%Y"
gw.states <- read_tsv(here("Data", "data_raw", "Gleditsch Ward", "iisystem.dat"),
col_names = c("gwcode", "cowc", "country.name",
"date.start", "date.end"),
col_types = cols(
gwcode = col_integer(),
cowc = col_character(),
country.name = col_character(),
date.start = col_date(format = gw.date),
date.end = col_date(format = gw.date)
),
locale = locale(encoding = "windows-1252"))
gw.microstates <- read_tsv(here("Data", "data_raw", "Gleditsch Ward", "microstatessystem.dat"),
col_names = c("gwcode", "cowc", "country.name",
"date.start", "date.end"),
col_types = cols(
gwcode = col_integer(),
cowc = col_character(),
country.name = col_character(),
date.start = col_date(format = gw.date),
date.end = col_date(format = gw.date)
),
locale = locale(encoding = "windows-1252"))
gw.codes <- bind_rows(gw.states, gw.microstates) %>%
filter(date.end > ymd("2000-01-01")) %>%
# Help out countrycode()
mutate(country.name = recode(country.name,
`Vietnam, Democratic Republic of` = "Vietnam",
`Yemen (Arab Republic of Yemen)` = "Yemen")) %>%
mutate(iso3 = countrycode(country.name, "country.name", "iso3c",
custom_match = c(Abkhazia = NA,
`South Ossetia` = NA,
Yugoslavia = "YUG",
Kosovo = "XKK",
`Korea, People's Republic of` = NA)),
iso2 = countrycode(iso3, "iso3c", "iso2c",
custom_match = c(XKK = "XK",
YUG = 345)),
cowcode = countrycode(iso3, "iso3c", "cown",
custom_match = c(XKK = 347,
SRB = 340,
YUG = 345,
VNM = 816))) %>%
filter(!is.na(iso3)) %>%
mutate(country.name = countrycode(iso3, "iso3c", "country.name",
custom_match = c(XKK = "Kosovo",
YUG = "Yugoslavia"))) %>%
select(country.name, iso3, iso2, gwcode, cowcode)
manual.gw <- tribble(
~country.name, ~iso3, ~iso2, ~gwcode, ~cowcode,
"Somaliland", "SOL", NA, 521, 521,
"Palestine (West Bank)", "PWB", NA, 667, 667,
"Palestine (Gaza)", "PGZ", NA, 668, 668,
"Palestine", "PSE", "PS", 669, 669,
"Hong Kong", "HKG", "HK", 715, 715
)
gw.codes <- bind_rows(gw.codes, manual.gw) %>%
arrange(country.name)
gw.codes %>% datatable()Foreign aid
OECD and AidData
The OECD collects detailed data on all foreign aid flows (ODA) from OECD member countries (and some non-member countries), mulilateral organizations, and the Bill and Melinda Gates Foundation (for some reason they’re the only nonprofit donor) to all DAC-eligible countries (and some non non-DAC-eligible countries).
The OECD tracks all this in a centralized Creditor Reporting System database and provides a nice front end for it at OECD.Stat with an open (but inscrutable) API (raw CRS data is also available). There are a set of pre-built queries with information about ODA flows by donor, recipient, and sector (purpose), but the pre-built data sources do not include all dimensions of the data. For example, Table DAC2a includes columns for donor, recipient, year, and total ODA (e.g. the US gave $X to Nigeria in 2008) , but does not indicate the purpose/sector for the ODA. Table DAC5 includes columns for the donor, sector, year, and total ODA (e.g. the US gave $X for education in 2008), but does not include recipient information.
Instead of using these pre-built queries or attempting to manipulate their parameters, it’s possible to use the OECD’s QWIDS query builder to create a custom download of data. However, it is slow and clunky and requires singificant munging and filtering after exporting.
The solution to all of this is to use data from AidData, which imports raw data from the OECD, cleans it, verifies it, and makes it freely available on GitHub.
AidData offers multiple versions of the data, including a full release, a thin release, aggregated donor/recipient/year data, and aggregated donor/recipient/year/purpose data. For the purposes of this study, all we care about are ODA flows by donor, recipient, year, and purpose, which is one of the ready-made datasets.
Notably, this aggregated data shows total aid commitments, not aid disbursements. Both types of ODA information are available from the OECD and it’s possible to get them using OECD’s raw data. However, AidData notes that disbursment data is sticky and slow—projects take a long time to fulfil and actual inflows of aid in a year can be tied to commitments made years before. Because we’re interested in donor reactions to restrictions on NGOs, any reaction would be visible in the decision to commit money to aid, not in the ultimate disbursement of aid, which is most likely already legally obligated and allocated to the country regardless of restrictions.
So, we look at ODA commitments.
aiddata.url <- "https://github.com/AidData-WM/public_datasets/releases/download/v3.1/AidDataCore_ResearchRelease_Level1_v3.1.zip"
aiddata.path <- here("Data", "data_raw", "AidData")
aiddata.zip.name <- basename(aiddata.url)
aiddata.name <- tools::file_path_sans_ext(aiddata.zip.name)
aiddata.final.name <- "AidDataCoreDonorRecipientYearPurpose_ResearchRelease_Level1_v3.1.csv"
# Download AidData data if needed
if (!file.exists(file.path(aiddata.path, aiddata.final.name))) {
aiddata.get <- GET(aiddata.url,
write_disk(file.path(aiddata.path, aiddata.zip.name),
overwrite = TRUE),
progress())
unzip(file.path(aiddata.path, aiddata.zip.name), exdir = aiddata.path)
# Clean up zip file and unnecessary CSV files
file.remove(file.path(aiddata.path, aiddata.zip.name))
list.files(aiddata.path, pattern = "csv", full.names = TRUE) %>%
map(~ ifelse(str_detect(.x, "DonorRecipientYearPurpose"), 0,
file.remove(file.path(.x))))
}
# Clean up AidData data
aiddata.raw <- read_csv(file.path(aiddata.path, aiddata.final.name),
col_types = cols(
donor = col_character(),
recipient = col_character(),
year = col_integer(),
coalesced_purpose_code = col_double(),
coalesced_purpose_name = col_character(),
commitment_amount_usd_constant_sum = col_double()
))
aiddata.clean <- aiddata.raw %>%
# Get rid of non-country recipients
filter(!str_detect(recipient,
regex("regional|unspecified|multi|value|global|commission",
ignore_case = TRUE))) %>%
filter(year < 9999) %>%
mutate(purpose.code.short = as.integer(str_sub(coalesced_purpose_code, 1, 3)))
# Donor, recipient, and purpose details
# I pulled these country names out of the dropdown menu at OECD.Stat Table 2a online
dac.donors <- c("Australia", "Austria", "Belgium", "Canada", "Czech Republic",
"Denmark", "Finland", "France", "Germany", "Greece", "Iceland",
"Ireland", "Italy", "Japan", "Korea", "Luxembourg", "Netherlands",
"New Zealand", "Norway", "Poland", "Portugal", "Slovak Republic",
"Slovenia", "Spain", "Sweden", "Switzerland", "United Kingdom",
"United States")
non.dac.donors <- c("Bulgaria", "Croatia", "Cyprus", "Estonia", "Hungary",
"Israel", "Kazakhstan", "Kuwait", "Latvia", "Liechtenstein",
"Lithuania", "Malta", "Romania", "Russia", "Saudi Arabia",
"Chinese Taipei", "Thailand", "Timor Leste", "Turkey",
"United Arab Emirates")
other.countries <- c("Brazil", "Chile", "Colombia", "India", "Monaco", "Qatar",
"South Africa", "Taiwan")
donors.all <- aiddata.clean %>%
distinct(donor) %>%
mutate(donor.type = case_when(
.$donor %in% c(dac.donors, non.dac.donors, other.countries) ~ "Country",
.$donor == "Bill & Melinda Gates Foundation" ~ "Private donor",
TRUE ~ "Multilateral or IGO"
))
donor.countries <- donors.all %>%
filter(donor.type == "Country") %>%
mutate(donor.cowcode = countrycode(donor, "country.name", "cown"),
donor.iso3 = countrycode(donor, "country.name", "iso3c"))
donors <- bind_rows(filter(donors.all, donor.type != "Country"),
donor.countries)
recipients <- aiddata.clean %>%
distinct(recipient) %>%
mutate(iso3 = countrycode(recipient, "country.name", "iso3c",
custom_match = c(`Korea, Democratic Republic of` = NA,
`Netherlands Antilles` = NA,
Kosovo = "XKK",
`Serbia and Montenegro` = "SCG",
Yugoslavia = "YUG"
))) %>%
left_join(gw.codes, by = "iso3") %>%
# Get rid of tiny countries
filter(!is.na(gwcode))
# Purposes
purposes <- aiddata.clean %>%
count(coalesced_purpose_name, coalesced_purpose_code)
purposes.url <- "https://www.oecd.org/dac/stats/documentupload/DAC_codeLists.xml"
purpose.nodes <- read_xml(purposes.url) %>% xml_find_all("//codelist-item")
purpose.codes <- data_frame(
code = purpose.nodes %>% xml_find_first(".//code") %>% xml_text(),
category = purpose.nodes %>% xml_find_first(".//category") %>% xml_text(),
name = purpose.nodes %>% xml_find_first(".//name") %>% xml_text(),
description = purpose.nodes %>% xml_find_first(".//description") %>% xml_text()
) %>%
mutate(code = as.integer(code))
# Extract the general categories of aid purposes (i.e. the first three digits of the purpose codes)
general.codes <- purpose.codes %>%
filter(code %in% as.character(100:1000) & str_detect(name, "^\\d")) %>%
mutate(code = as.integer(code)) %>%
select(purpose.code.short = code, purpose.category.name = name) %>%
mutate(purpose.category.clean = str_replace(purpose.category.name,
"\\d\\.\\d ", "")) %>%
separate(purpose.category.clean,
into = c("purpose.sector", "purpose.category"),
sep = ", ") %>%
mutate_at(vars(c(purpose.sector, purpose.category)), funs(str_to_title(.))) %>%
select(-purpose.category.name)
# These 7 codes are weird and get filtered out inadvertantly
codes.not.in.oecd.list <- tribble(
~purpose.code.short, ~purpose.sector, ~purpose.category,
100, "Social", "Social Infrastructure",
200, "Eco", "Economic Infrastructure",
300, "Prod", "Production",
310, "Prod", "Agriculture",
320, "Prod", "Industry",
420, "Multisector", "Women in development",
# NB: This actually is splict between 92010 (domestic NGOs), 92020
# (international NGOs), and 92030 (local and regional NGOs)
920, "Non Sector", "Support to NGOs"
)
purpose.codes.clean <- general.codes %>%
bind_rows(codes.not.in.oecd.list) %>%
arrange(purpose.code.short) %>%
mutate(purpose.contentiousness = "")
# Manually code contentiousness of purposes
write_csv(purpose.codes.clean,
here("Data", "data_manual",
"purpose_codes_contention_WILL_BE_OVERWRITTEN.csv"))
purpose.codes.contentiousness <- read_csv(here("Data", "data_manual",
"purpose_codes_contention.csv"))
aiddata.final <- aiddata.clean %>%
left_join(donors, by = "donor") %>%
left_join(recipients, by = "recipient") %>%
left_join(purpose.codes.contentiousness, by = "purpose.code.short") %>%
mutate(donor.type.collapsed = ifelse(donor.type == "Country", "Country",
"IGO, Multilateral, or Private")) %>%
select(donor, donor.type, donor.type.collapsed,
donor.cowcode, donor.iso3, year,
country.name, cowcode, gwcode, iso2, iso3,
oda = commitment_amount_usd_constant_sum,
purpose.code.short, purpose.sector, purpose.category,
purpose.contentiousness,
coalesced_purpose_code, coalesced_purpose_name) %>%
arrange(cowcode, year)
ever.dac.eligible <- read_csv(here("Data", "data_manual",
"oecd_dac_countries.csv")) %>%
# Ignore High Income Countries and More Advanced Developing Countries
filter(!(dac_abbr %in% c("HIC", "ADC"))) %>%
left_join(gw.codes, by = "iso3") %>%
select(cowcode) %>% unlist() %>% c() %>% unique()List of donors
List of recipients
List of purposes
Summary of clean data
## Observations: 624,258
## Variables: 18
## $ donor <chr> "Islamic Development Bank (ISDB)", "Islamic Development Bank (ISDB)", "Islamic Deve...
## $ donor.type <chr> "Multilateral or IGO", "Multilateral or IGO", "Multilateral or IGO", "Multilateral ...
## $ donor.type.collapsed <chr> "IGO, Multilateral, or Private", "IGO, Multilateral, or Private", "IGO, Multilatera...
## $ donor.cowcode <int> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,...
## $ donor.iso3 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,...
## $ year <int> 1982, 1983, 1985, 1988, 1989, 1989, 1990, 1992, 1992, 1993, 1995, 1995, 1995, 1995,...
## $ country.name <chr> "United States", "United States", "United States", "United States", "United States"...
## $ cowcode <dbl> 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,...
## $ gwcode <dbl> 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,...
## $ iso2 <chr> "US", "US", "US", "US", "US", "US", "US", "US", "US", "US", "US", "US", "US", "US",...
## $ iso3 <chr> "USA", "USA", "USA", "USA", "USA", "USA", "USA", "USA", "USA", "USA", "USA", "USA",...
## $ oda <dbl> 264104, 2657078, 2697117, 585643, 538475, 87616, 163344, 598636, 209523, 968333, 13...
## $ purpose.code.short <int> 111, 114, 160, 111, 111, 160, 111, 111, 160, 111, 111, 112, 113, 160, 998, 111, 113...
## $ purpose.sector <chr> "Social", "Social", "Social", "Social", "Social", "Social", "Social", "Social", "So...
## $ purpose.category <chr> "Education", "Education", "Other", "Education", "Education", "Other", "Education", ...
## $ purpose.contentiousness <chr> "Low", "Low", "Low", "Low", "Low", "Low", "Low", "Low", "Low", "Low", "Low", "Low",...
## $ coalesced_purpose_code <dbl> 11120, 11420, 16010, 11120, 11120, 16010, 11120, 11120, 16010, 11105, 11120, 11220,...
## $ coalesced_purpose_name <chr> "Education facilities and training", "Higher education", "Social/ welfare services"...USAID
USAID provides the complete dataset for its Foreign Aid Explorer as a giant CSV file. The data includes both economic and military aid, but it’s easy to filter out the military aid. Here we only look at obligations, not disbursements, so that the data is comparable to the OECD data from AidData. The data we downloaded provides constant amounts in 2015 dollars; we rescale that to 2011 to match all other variables.
usaid.url <- "https://explorer.usaid.gov/prepared/us_foreign_aid_complete.csv"
usaid.path <- here("Data", "data_raw", "USAID")
usaid.name <- basename(usaid.url)
# Download USAID data if needed
if (!file.exists(file.path(usaid.path, usaid.name))) {
usaid.get <- GET(usaid.url,
write_disk(file.path(usaid.path, usaid.name),
overwrite = TRUE),
progress())
}
# Clean up USAID data
usaid.raw <- read_csv(file.path(usaid.path, usaid.name),
na = c("", "NA", "NULL"),
col_types = cols(
country_id = col_integer(),
country_code = col_character(),
country_name = col_character(),
region_id = col_integer(),
region_name = col_character(),
income_group_id = col_integer(),
income_group_name = col_character(),
income_group_acronym = col_character(),
implementing_agency_id = col_integer(),
implementing_agency_acronym = col_character(),
implementing_agency_name = col_character(),
implementing_subagency_id = col_integer(),
subagency_acronym = col_character(),
subagency_name = col_character(),
channel_category_id = col_integer(),
channel_category_name = col_character(),
channel_subcategory_id = col_integer(),
channel_subcategory_name = col_character(),
channel_id = col_integer(),
channel_name = col_character(),
dac_category_id = col_integer(),
dac_category_name = col_character(),
dac_sector_code = col_integer(),
dac_sector_name = col_character(),
dac_purpose_code = col_integer(),
dac_purpose_name = col_character(),
funding_account_id = col_character(),
funding_account_name = col_character(),
funding_agency_id = col_integer(),
funding_agency_name = col_character(),
funding_agency_acronym = col_character(),
assistance_category_id = col_integer(),
assistance_category_name = col_character(),
aid_type_group_id = col_integer(),
aid_type_group_name = col_character(),
activity_id = col_integer(),
activity_name = col_character(),
activity_project_number = col_character(),
activity_start_date = col_date(format = ""),
activity_end_date = col_date(format = ""),
transaction_type_id = col_integer(),
transaction_type_name = col_character(),
fiscal_year = col_character(),
current_amount = col_double(),
constant_amount = col_double(),
USG_sector_id = col_integer(),
USG_sector_name = col_character(),
submission_id = col_integer(),
numeric_year = col_double()
))
usaid.clean <- usaid.raw %>%
filter(assistance_category_name == "Economic") %>%
filter(transaction_type_name == "Obligations") %>%
mutate(country_code = recode(country_code, `CS-KM` = "XKK")) %>%
# Remove regions and World
filter(!str_detect(country_name, "Region")) %>%
filter(!(country_name %in% c("World"))) %>%
left_join(gw.codes, by = c("country_code" = "iso3")) %>%
# Get rid of tiny and old states
filter(!is.na(cowcode)) %>%
select(cowcode, year = numeric_year,
implementing_agency_name, subagency_name, activity_name,
channel_category_name, channel_subcategory_name, dac_sector_code,
oda.us.current = current_amount, oda.us.2015 = constant_amount) %>%
mutate(aid.deflator = oda.us.current / oda.us.2015 * 100) %>%
mutate(channel.ngo.us = channel_subcategory_name == "NGO - United States",
channel.ngo.int = channel_subcategory_name == "NGO - International",
channel.ngo.dom = channel_subcategory_name == "NGO - Non United States")
# Get rid of this because it's huge and taking up lots of memory
rm(usaid.raw)Implementing agencies
Here are the US government agencies giving out money:
Activities
The activities listed don’t follow any standard coding guidelines. There are tens of thousands of them. Here are the first 100, just for reference:
Channels
USAID distinguishes between domestic, foreign, and international NGOs, companies, multilateral organizations, etc. recipients (or channels) of money:
Summary of clean data
## Observations: 324,559
## Variables: 14
## $ cowcode <dbl> 200, 666, 666, 666, 666, 666, 666, 645, 200, 666, 666, 666, 666, 666, 666, 666, 66...
## $ year <dbl> 1947, 1985, 1985, 1986, 1986, 1991, 1991, 2004, 1949, 1994, 1992, 1989, 1998, 1993...
## $ implementing_agency_name <chr> "Department of the Treasury", "U.S. Agency for International Development", "U.S. A...
## $ subagency_name <chr> "not applicable", "not applicable", "not applicable", "not applicable", "not appli...
## $ activity_name <chr> "British Loan", "USAID Grants", "ESF", "ESF", "USAID Grants", "ESF", "USAID Grants...
## $ channel_category_name <chr> "Government", "Government", "Government", "Government", "Government", "Government"...
## $ channel_subcategory_name <chr> "Government - United States", "Government - United States", "Government - United S...
## $ dac_sector_code <int> 430, 430, 430, 430, 430, 430, 430, 210, 430, 430, 430, 430, 430, 430, 430, 430, 43...
## $ oda.us.current <dbl> 3750000000, 1950050000, 1950050000, 1898400000, 1898400000, 1850000000, 1850000000...
## $ oda.us.2015 <dbl> 33891858552, 3761152948, 3761152948, 3579858613, 3579858613, 2962911687, 296291168...
## $ aid.deflator <dbl> 11.06460, 51.84713, 51.84713, 53.03003, 53.03003, 62.43858, 62.43858, 80.55505, 12...
## $ channel.ngo.us <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE...
## $ channel.ngo.int <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE...
## $ channel.ngo.dom <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE...NGO regulations
NGO legislation
In 2013, Darin Christensen and Jeremy Weinstein collected detailed data on NGO regulations for their Journal of Democracy article. Suparna Chaudhry expanded this data as part of her dissertation research, but wants her data embargoed until some of her work is publsihed. To prevent data leakage, the code to make minor manual adjustments is included in an untracked file and not made public.
The code below will still work on the original Christensen and Weinstein data, but it will (obviously) not include Chaudhry’s expanded data.
# Original DCJW data
dcjw.orig.path <- here("Data", "data_raw", "DCJW NGO Laws", "DCJW_NGO_Laws.xlsx")
dcjw.orig <- read_excel(dcjw.orig.path)[,1:50] %>%
select(-c(dplyr::contains("source"), dplyr::contains("burden"),
dplyr::contains("subset"), Coder, Date))
# Tidy DCJW data
dcjw.path <- here("Data", "data_raw", "DCJW NGO Laws", "Admn Crackdown_updated.xlsx")
dcjw.questions.raw <- read_csv(here("Data", "data_manual", "dcjw_questions.csv"))
dcjw.barriers.clean <- dcjw.questions.raw %>%
distinct(question_cat, barrier)
dcjw.barriers.ignore <- dcjw.questions.raw %>%
select(question, ignore_in_index)
dcjw <- read_excel(dcjw.path)[,1:50] %>%
select(-c(dplyr::contains("source"), dplyr::contains("burden"),
dplyr::contains("subset"), Coder, Date)) %>%
gather(key, value, -Country) %>%
separate(key, c("question", "var.name"), 4) %>%
filter(!is.na(Country)) %>%
mutate(var.name = ifelse(var.name == "", "value", gsub("_", "", var.name))) %>%
spread(var.name, value) %>%
# Get rid of rows where year is missing and regulation was not imposed
filter(!(is.na(year) & value == 0)) %>%
# Some entries have multiple years; for now just use the first year
mutate(year = str_split(year, ",")) %>% unnest(year) %>%
group_by(Country, question) %>% slice(1) %>% ungroup() %>%
mutate(value = as.integer(value), year = as.integer(year)) %>%
# If year is missing but some regulation exists, assume it has always already
# existed (since 1950, arbitrarily)
mutate(year = ifelse(is.na(year), 1950, year))
potential.dcjw.panel.question <- dcjw %>%
tidyr::expand(Country, question,
year = min(.$year, na.rm = TRUE):2015)
dcjw.panel.all.laws <- dcjw %>%
right_join(potential.dcjw.panel.question,
by = c("Country", "question", "year"))
# Suparna Chaudry updated the original DCJW data, but wants the data embargoed
# until some of her dissertation is published. To prevent data leakage onto the
# internet, minor manual adjustments are included in a hidden file here instead
# of this main file.
source(here("Data", "data_raw", "DCJW NGO Laws", "chaudhry_manual_changes.R"))
if (all.na.means.zero) {
dcjw.panel.all.laws <- dcjw.panel.all.laws %>%
group_by(Country, question) %>%
# Bring most recent legislation forward
mutate(value = zoo::na.locf(value, na.rm = FALSE)) %>%
ungroup() %>%
# Set defaults for columns that aren't all NA
# I could be fancy and consolidate these conditionals into just one, but this
# gives us more flexibility to set different defaults per law (like how q_2b
# or q_4c might be better as NA_integer_ instead of 0)
mutate(value.fixed = case_when(
is.na(.$value) & .$question == "q_1a" ~ 0L,
is.na(.$value) & .$question == "q_1b" ~ 0L,
is.na(.$value) & .$question == "q_2a" ~ 0L,
is.na(.$value) & .$question == "q_2b" ~ 0L,
is.na(.$value) & .$question == "q_2c" ~ 0L,
is.na(.$value) & .$question == "q_2d" ~ 0L,
is.na(.$value) & .$question == "q_3a" ~ 0L,
is.na(.$value) & .$question == "q_3b" ~ 0L,
is.na(.$value) & .$question == "q_3c" ~ 0L,
is.na(.$value) & .$question == "q_3d" ~ 0L,
is.na(.$value) & .$question == "q_3e" ~ 0L,
is.na(.$value) & .$question == "q_3f" ~ 0L,
is.na(.$value) & .$question == "q_4a" ~ 0L,
is.na(.$value) & .$question == "q_4b" ~ 0L,
is.na(.$value) & .$question == "q_4c" ~ 0L,
TRUE ~ .$value
)) %>%
select(-value) %>%
spread(question, value.fixed) %>%
ungroup()
} else {
dcjw.panel.all.laws <- dcjw.panel.all.laws %>%
group_by(Country, question) %>%
# Bring most recent legislation forward
mutate(value = zoo::na.locf(value, na.rm = FALSE)) %>%
mutate(not.all.na = !all(is.na(value))) %>%
ungroup() %>%
# Set defaults for columns that aren't all NA
# I could be fancy and consolidate these conditionals into just one, but this
# gives us more flexibility to set different defaults per law (like how q_2b
# or q_4c might be better as NA_integer_ instead of 0)
mutate(value.fixed = case_when(
.$not.all.na & is.na(.$value) & .$question == "q_1a" ~ 0L,
.$not.all.na & is.na(.$value) & .$question == "q_1b" ~ 0L,
.$not.all.na & is.na(.$value) & .$question == "q_2a" ~ 0L,
.$not.all.na & is.na(.$value) & .$question == "q_2b" ~ 0L,
.$not.all.na & is.na(.$value) & .$question == "q_2c" ~ 0L,
.$not.all.na & is.na(.$value) & .$question == "q_2d" ~ 0L,
.$not.all.na & is.na(.$value) & .$question == "q_3a" ~ 0L,
.$not.all.na & is.na(.$value) & .$question == "q_3b" ~ 0L,
.$not.all.na & is.na(.$value) & .$question == "q_3c" ~ 0L,
.$not.all.na & is.na(.$value) & .$question == "q_3d" ~ 0L,
.$not.all.na & is.na(.$value) & .$question == "q_3e" ~ 0L,
.$not.all.na & is.na(.$value) & .$question == "q_3f" ~ 0L,
.$not.all.na & is.na(.$value) & .$question == "q_4a" ~ 0L,
.$not.all.na & is.na(.$value) & .$question == "q_4b" ~ 0L,
.$not.all.na & is.na(.$value) & .$question == "q_4c" ~ 0L,
TRUE ~ .$value
)) %>%
select(-not.all.na, -value) %>%
spread(question, value.fixed) %>%
ungroup()
}
potential.dcjw.panel.barrier <- potential.dcjw.panel.question %>%
mutate(question_cat = as.integer(substr(question, 3, 3))) %>%
left_join(dcjw.barriers.clean, by = "question_cat") %>%
distinct(Country, year, barrier)
dcjw.panel.barriers <- dcjw.panel.all.laws %>%
gather(question, value, -Country, -year) %>%
mutate(question_cat = as.integer(substr(question, 3, 3))) %>%
left_join(dcjw.barriers.clean, by = "question_cat") %>%
# Make an index for each type of barrier
mutate(value = as.numeric(value)) %>%
mutate(value = case_when(
# Reverse values for associational rights
.$question == "q_1a" & .$value == 0 ~ 1,
.$question == "q_1a" & .$value == 1 ~ 0,
.$question == "q_1b" & .$value == 0 ~ 1,
.$question == "q_1b" & .$value == 1 ~ 0,
# Reverse value for q_2c
.$question == "q_2c" & .$value == 0 ~ 1,
.$question == "q_2c" & .$value == 1 ~ 0,
# Recode 0-2 questions as 0-1
.$question == "q_3e" & .$value == 1 ~ 0.5,
.$question == "q_3e" & .$value == 2 ~ 1,
.$question == "q_3f" & .$value == 1 ~ 0.5,
.$question == "q_3f" & .$value == 2 ~ 1,
.$question == "q_4a" & .$value == 1 ~ 0.5,
.$question == "q_4a" & .$value == 2 ~ 1,
TRUE ~ .$value
)) %>%
# Ignore neutral indexes, like basic registration requirements
left_join(dcjw.barriers.ignore, by = "question") %>%
mutate(value_restrictive = ifelse(ignore_in_index, 0, value)) %>%
group_by(Country, year, barrier) %>%
# Add up all the barriers for each country year
# Use a floor of zero to account for negative values
summarise(all = sum(value, na.rm = TRUE),
restrictive = sum(value_restrictive, na.rm = TRUE)) %>%
mutate_at(vars(all, restrictive),
funs(ifelse(. < 0, as.integer(0), .))) %>%
# Join with full possible panel
right_join(potential.dcjw.panel.barrier,
by = c("Country", "barrier", "year")) %>%
gather(temp, value, all, restrictive) %>%
unite(temp1, barrier, temp, sep = "_") %>%
spread(temp1, value) %>%
ungroup() %>%
# Take "_restrictive" out of the variable names.
# "entry" = all restrictive entry laws, "entry_all" = all entry laws
rename_(.dots = setNames(paste0(unique(dcjw.questions.raw$barrier), "_restrictive"),
unique(dcjw.questions.raw$barrier))) %>%
# Standardize barrier indexes by dividing by maximum number possible
mutate_at(vars(entry, funding, advocacy),
funs(std = . / max(.))) %>%
mutate(barriers.total = advocacy + entry + funding,
barriers.total_std = advocacy_std + entry_std + funding_std)
dcjw.full <- dcjw.panel.all.laws %>%
left_join(dcjw.panel.barriers, by = c("Country", "year")) %>%
# Lop off the ancient observations
filter(year > 1980) %>%
# Rename q_* variables
rename_(.dots = setNames(dcjw.questions.raw$question,
dcjw.questions.raw$question_clean)) %>%
# Add additional variables
mutate(Country = countrycode(Country, "country.name", "country.name"),
cowcode = countrycode(Country, "country.name", "cown",
custom_match = c(Serbia = 340,
Vietnam = 816))) %>%
select(country.name = Country, cowcode, year, everything())Chaudhry’s data augments Christensen and Weinstein’s data substantially. Christensen and Weinstein include information for 98 countries, while Chaudhry includes 148 countries.
In addition to panel data on the presence or absence of specific NGO regulations, we create several indexes for each of the categories of regulation, following Christensen and Weinstein’s classification:
entry(Q2b, Q2c, Q2d; 3 points maximum, actual max = 3 points maximum): barriers to entry- Q2c is reversed, so not being allowed to appeal registration status earns 1 point.
- Q2a is omitted because it’s benign
funding(Q3b, Q3c, Q3d, Q3e, Q3f; 5 points maximum, actual max = 4.5): barriers to funding- Q3a is omitted because it’s benign
- Scores that range between 0–2 are rescaled to 0–1 (so 1 becomes 0.5)
advocacy(Q4a, Q4c; 2 points maximum, actual max = 2): barriers to advocacy- Q4b is omitted because it’s not a law
- Scores that range between 0–2 are rescaled to 0–1 (so 1 becomes 0.5)
barriers.total(10 points maximum, actual max = 8.5): sum of all three indexes
These indexes are also standardized by dividing by the maximum, yielding the following variables:
entry_std: 1 point maximum, actual max = 1funding_std: 1 point maximum, actual max = 1advocacy_std: 1 point maximum, actual max = 1barriers.total_std: 3 points maximum, actual max = 2.5
## Observations: 5,180
## Variables: 31
## $ country.name <chr> "Afghanistan", "Afghanistan", "Afghanistan", "Afghanistan", "Afghanistan", "...
## $ cowcode <dbl> 700, 700, 700, 700, 700, 700, 700, 700, 700, 700, 700, 700, 700, 700, 700, 7...
## $ year <dbl> 1981, 1982, 1983, 1984, 1985, 1986, 1987, 1988, 1989, 1990, 1991, 1992, 1993...
## $ const_assoc <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1...
## $ political_parties <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1...
## $ ngo_register <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1...
## $ ngo_register_burden <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1...
## $ ngo_register_appeal <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1...
## $ ngo_barrier_foreign_funds <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1...
## $ ngo_disclose_funds <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1...
## $ ngo_foreign_fund_approval <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0...
## $ ngo_foreign_fund_channel <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0...
## $ ngo_foreign_fund_restrict <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0...
## $ ngo_foreign_fund_prohibit <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0...
## $ ngo_type_foreign_fund_prohibit <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0...
## $ ngo_politics <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0...
## $ ngo_politics_intimidation <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0...
## $ ngo_politics_foreign_fund <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0...
## $ advocacy_all <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0...
## $ advocacy <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0...
## $ association_all <dbl> 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 0, 0, 0...
## $ association <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0...
## $ entry_all <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 3, 3...
## $ entry <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2...
## $ funding_all <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1...
## $ funding <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0...
## $ entry_std <dbl> 0.3333333, 0.3333333, 0.3333333, 0.3333333, 0.3333333, 0.3333333, 0.3333333,...
## $ funding_std <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0...
## $ advocacy_std <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0...
## $ barriers.total <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2...
## $ barriers.total_std <dbl> 0.3333333, 0.3333333, 0.3333333, 0.3333333, 0.3333333, 0.3333333, 0.3333333,...Civil society regulatory environment
An alternative way of measuring civil society restrictions is to look at the overal civil society regulatory environment rather than specific laws, since de jure restrictions do not always map clearly into de facto restrictions (especially in dictatorships where the implementation of laws is more discretionary).
Andrew Heiss develops a new civil society regulatory environment index (CSRE) in his dissertation, which combines two civil society indexes from the Varieties of Democracy project (V-Dem): (1) civil society repression and (2) civil society entry and exit regulations. The CSRE ranges from roughly −6 to 6 (though typically only from −4 to 4ish), and shows more variation over time since it ostensibly captures changes in the implementation of the regulatory environment rather than the presence or absence of legislation.
While the main focus of this paper is donor response to new legislation, we also look at donor response to changes in the overall CSRE as a robustness check.
There’s no direct link for V-Dem data, since it’s behind a contact information form. So, download the 8.0 version of the “Country-Year: V-Dem + other” dataset and place it in Data/data_raw/V-Dem/v8
When loading the CSV, readr::read_csv() chokes on a bunch of rows for whatever reason and gives a ton of warnings, but it still works. Loading the Stata version of V-Dem doesn’t create the warnings, but it’s slower and it results in the same data. So we just load from CSV and make sure it has the right number of rows and columns in the end.
vdem.raw <- read_csv(here("Data", "data_raw", "V-Dem", "v8",
"V-Dem-CY+Others-v8.csv"))
testthat::expect_equal(nrow(vdem.raw), 26537)
testthat::expect_equal(ncol(vdem.raw), 4641)# Extract civil society-related variables and create CSRE index
vdem.cso <- vdem.raw %>%
# Missing COW codes
mutate(COWcode = case_when(
.$country_text_id == "SML" ~ 521L,
.$country_text_id == "PSE" ~ 667L,
.$country_text_id == "PSG" ~ 668L,
TRUE ~ .$COWcode)) %>%
select(country_name, year, cowcode = COWcode,
e_fh_ipolity2, v2x_corr, e_wbgi_gee,
v2x_frassoc_thick, v2xcs_ccsi,
starts_with("v2cseeorgs"),
starts_with("v2csreprss"),
starts_with("v2cscnsult"),
starts_with("v2csprtcpt"),
starts_with("v2csgender"),
starts_with("v2csantimv")) %>%
mutate(v2csreprss_ord = factor(v2csreprss_ord,
labels = c("Severely", "Substantially",
"Moderately", "Weakly", "No"),
ordered = TRUE),
csre = v2csreprss + v2cseeorgs) %>%
filter(year > 1980)Civil society restrictions data
Summary of clean data
## Observations: 6,328
## Variables: 30
## $ country_name <chr> "United States of America", "United States of America", "United States of America",...
## $ year <int> 1981, 1982, 1983, 1984, 1985, 1986, 1987, 1988, 1989, 1990, 1991, 1992, 1993, 1994,...
## $ cowcode <int> 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,...
## $ csre <dbl> 5.532625, 5.532625, 5.532625, 5.532625, 5.532625, 5.532625, 5.532625, 5.532625, 5.5...
## $ v2csreprss_ord <ord> No, No, No, No, No, No, No, No, No, No, No, No, No, No, No, No, No, No, No, No, No,...
## $ v2csreprss <dbl> 2.831333, 2.831333, 2.831333, 2.831333, 2.831333, 2.831333, 2.831333, 2.831333, 2.8...
## $ v2csreprss_codehigh <dbl> 3.350354, 3.350354, 3.350354, 3.350354, 3.350354, 3.350354, 3.350354, 3.350354, 3.3...
## $ v2csreprss_codelow <dbl> 1.970434, 1.970434, 1.970434, 1.970434, 1.970434, 1.970434, 1.970434, 1.970434, 1.9...
## $ v2csreprss_mean <dbl> 4.000000, 4.000000, 4.000000, 4.000000, 4.000000, 4.000000, 4.000000, 4.000000, 4.0...
## $ v2csreprss_nr <int> 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 15, 15, 15,...
## $ v2csreprss_ord_codehigh <int> 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4,...
## $ v2csreprss_ord_codelow <int> 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4,...
## $ v2csreprss_osp <dbl> 3.912476, 3.912476, 3.912476, 3.912476, 3.912476, 3.912476, 3.912476, 3.912476, 3.9...
## $ v2csreprss_osp_codehigh <dbl> 3.999951, 3.999951, 3.999951, 3.999951, 3.999951, 3.999951, 3.999951, 3.999951, 3.9...
## $ v2csreprss_osp_codelow <dbl> 3.846990, 3.846990, 3.846990, 3.846990, 3.846990, 3.846990, 3.846990, 3.846990, 3.8...
## $ v2csreprss_osp_sd <dbl> 0.1378864, 0.1378864, 0.1378864, 0.1378864, 0.1378864, 0.1378864, 0.1378864, 0.1378...
## $ v2csreprss_sd <dbl> 0.6943592, 0.6943592, 0.6943592, 0.6943592, 0.6943592, 0.6943592, 0.6943592, 0.6943...
## $ v2cseeorgs <dbl> 2.701292, 2.701292, 2.701292, 2.701292, 2.701292, 2.701292, 2.701292, 2.701292, 2.7...
## $ v2cseeorgs_codehigh <dbl> 3.212703, 3.212703, 3.212703, 3.212703, 3.212703, 3.212703, 3.212703, 3.212703, 3.2...
## $ v2cseeorgs_codelow <dbl> 2.277519, 2.277519, 2.277519, 2.277519, 2.277519, 2.277519, 2.277519, 2.277519, 2.2...
## $ v2cseeorgs_mean <dbl> 3.625000, 3.625000, 3.625000, 3.625000, 3.625000, 3.625000, 3.625000, 3.625000, 3.6...
## $ v2cseeorgs_nr <int> 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 8, 15, 15, 15,...
## $ v2cseeorgs_ord <int> 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4,...
## $ v2cseeorgs_ord_codehigh <int> 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4,...
## $ v2cseeorgs_ord_codelow <int> 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4,...
## $ v2cseeorgs_osp <dbl> 3.780493, 3.780493, 3.780493, 3.780493, 3.780493, 3.780493, 3.780493, 3.780493, 3.7...
## $ v2cseeorgs_osp_codehigh <dbl> 3.955077, 3.955077, 3.955077, 3.955077, 3.955077, 3.955077, 3.955077, 3.955077, 3.9...
## $ v2cseeorgs_osp_codelow <dbl> 3.671487, 3.671487, 3.671487, 3.671487, 3.671487, 3.671487, 3.671487, 3.671487, 3.6...
## $ v2cseeorgs_osp_sd <dbl> 0.16074768, 0.16074768, 0.16074768, 0.16074768, 0.16074768, 0.16074768, 0.16074768,...
## $ v2cseeorgs_sd <dbl> 0.4812464, 0.4812464, 0.4812464, 0.4812464, 0.4812464, 0.4812464, 0.4812464, 0.4812...Neighboring states
Calculate three types of distance between all countries with the CShapes R package: mimimum distance, capital distance, and centroid distance. Because this takes a really long time, it’s best to run Data/get_distances.R separately elsewhere first (like on a VPS) and then save the resulting .rds files in Data/data_raw/Country Distances.
The all.distances data frame below contains all three distance types between every country and every other country in 2012 (though it could potentially include distances from 1991–2015). This is used later to weight variables by distance. It also includes an indicator variable called is.rough.neighbor that is true if the two countries are less than 900 km apart, which captures neighbor relationships better than simple contiguous borders.
distance.folder <- here("Data", "data_raw", "Country distances")
read_distances <- function(type, year) {
stopifnot(year >= 1980, year <= 2015)
stopifnot(type %in% c("capital", "cent", "min"))
# Read in the RDS file for the type and year
matrix.raw <- readRDS(file.path(distance.folder,
paste0(type, "_", year, "-01-01.rds")))
# Convert distance matrix to long dataframe
df.clean <- as.data.frame(matrix.raw) %>%
mutate(gwcode = rownames(.)) %>%
gather(gwcode.other, distance, -gwcode) %>%
# Set inverted diagonal 0s to 0, real 0s to 1
mutate(distance.inv = ifelse(gwcode == gwcode.other & distance == 0, 0,
ifelse(gwcode != gwcode.other & distance == 0, 1,
1 / distance))) %>%
mutate(type = type, year = year) %>%
mutate_at(vars(starts_with("gwcode")), funs(as.numeric)) %>%
# Mark if country is within 900 km
mutate(is.rough.neighbor = distance < 900) %>%
# Standardize inverted distances so that all rows within a country add to 1
# so that it can be used in weighted.mean.
# This is the same as calculating the row sum of the matrix
group_by(gwcode) %>%
mutate(distance.std = distance.inv / sum(distance.inv)) %>%
ungroup() %>%
left_join(gw.codes, by = "gwcode") %>%
left_join(gw.codes, by = c("gwcode.other" = "gwcode"),
suffix = c(".self", ".other"))
return(df.clean)
}
# Load and combine minimum distance matrices for all years (could potentially
# also do "cent" or "capital")
all.distances <- expand.grid(year = 1980:2015,
type = "min",
stringsAsFactors = FALSE) %>%
mutate(distances = map2(type, year, ~ read_distances(.x, .y))) %>%
unnest(distances)
all.distances %>% glimpse()## Observations: 1,226,302
## Variables: 18
## $ year <int> 1980, 1980, 1980, 1980, 1980, 1980, 1980, 1980, 1980, 1980, 1980, 1980, 1980, 1980, 1980...
## $ type <chr> "min", "min", "min", "min", "min", "min", "min", "min", "min", "min", "min", "min", "min...
## $ gwcode <dbl> 2, 20, 31, 40, 41, 42, 51, 52, 53, 54, 55, 56, 57, 70, 90, 91, 92, 93, 94, 95, 100, 101,...
## $ gwcode.other <dbl> 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2...
## $ distance <dbl> 0.0000, 0.0000, 105.6785, 233.2897, 931.8133, 1070.4367, 782.9170, 2544.8651, 2540.5566,...
## $ distance.inv <dbl> 0.0000000000, 1.0000000000, 0.0094626606, 0.0042865160, 0.0010731763, 0.0009341982, 0.00...
## $ type1 <chr> "min", "min", "min", "min", "min", "min", "min", "min", "min", "min", "min", "min", "min...
## $ year1 <int> 1980, 1980, 1980, 1980, 1980, 1980, 1980, 1980, 1980, 1980, 1980, 1980, 1980, 1980, 1980...
## $ is.rough.neighbor <lgl> TRUE, TRUE, TRUE, TRUE, FALSE, FALSE, TRUE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, TR...
## $ distance.std <dbl> 0.000000e+00, 9.684943e-01, 1.424432e-01, 5.810121e-02, 1.014570e-03, 8.934232e-04, 2.57...
## $ country.name.self <chr> "United States", "Canada", "Bahamas", "Cuba", "Haiti", "Dominican Republic", "Jamaica", ...
## $ iso3.self <chr> "USA", "CAN", "BHS", "CUB", "HTI", "DOM", "JAM", "TTO", "BRB", "DMA", "GRD", "LCA", "VCT...
## $ iso2.self <chr> "US", "CA", "BS", "CU", "HT", "DO", "JM", "TT", "BB", "DM", "GD", "LC", "VC", "MX", "GT"...
## $ cowcode.self <dbl> 2, 20, 31, 40, 41, 42, 51, 52, 53, 54, 55, 56, 57, 70, 90, 91, 92, 93, 94, 95, 100, 101,...
## $ country.name.other <chr> "United States", "United States", "United States", "United States", "United States", "Un...
## $ iso3.other <chr> "USA", "USA", "USA", "USA", "USA", "USA", "USA", "USA", "USA", "USA", "USA", "USA", "USA...
## $ iso2.other <chr> "US", "US", "US", "US", "US", "US", "US", "US", "US", "US", "US", "US", "US", "US", "US"...
## $ cowcode.other <dbl> 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2...Other controls and alternative hypotheses
There are a bunch of reasons why overall aid flows might change over time. We controle for these alternative hypotheses:
- Regime type: donors likely give less aid to democracies
- Polity IV:
polity(imputed 0-10 Polity score from V-Dem, originallye_fh_ipolity2) - Wealth: donors likely give less aid to richer countries
- GDP per capita:
gdp.capita(GDP per capita (constant 2011 USD) from WDI) - Trade as % of GDP:
trade.pct.gdp(from WDI) - Aid dependency:
aid.pct.gdp(Constant 2011 ODA as percent of constant 2011 GDP from AidData and WDI) - Government capacity: donors likely give more aid to NGOs in countries with weak institutions (and perhaps less aid overall?)
- Corruption:
corruption(0-10 scale from V-Dem: public sector + executive + + legislative + judicial corruption, originallyv2x_corron 0-1) - BAD VARIABLE, almost 50% missing; sadface Quality of governance:
govt.effectiveness(≈-2 to 2 scale from V-Dem and WBGI, originallye_wbgi_gee) - Bad stuff: donors likely give more aid to countries when bad things happen like wars and natural disasters
- Internal conflict:
internal.conflict.past.5(binary indicator variable if there was a conflict in that year or in the previous 5 years from UCDP/PRIO) - Disasters:
natural_disaster.affected,natural_disaster.deaths,natural_disaster.occurrence, andnatural_disaster.total.affected, among others (from EM-DAT)
World Bank and UN development indicators
The World Bank has a fantastic API for their statistical indicators and there’s already a nice R package for accessing it, so downloading indicators is trivial. For the sake of our models, we care about GDP per capita as a proxy for a country’s overall development. We also collect a few other relevant variables just for fun.
# UN country codes
un.codes <- read_csv(here("Data", "data_manual", "un_codes.csv")) %>%
select(un_code, iso3)
# GDP by Type of Expenditure at constant (2005) prices - US dollars
# http://data.un.org/Data.aspx?q=gdp&d=SNAAMA&f=grID%3a102%3bcurrID%3aUSD%3bpcFlag%3a0
un.gdp.raw <- read_csv(here("Data", "data_raw", "UN data",
"UNdata_Export_20170125_042818046.csv")) %>%
rename(un_code = `Country or Area Code`) %>%
mutate(value.type = "Constant")
# http://data.un.org/Data.aspx?q=gdp&d=SNAAMA&f=grID%3a101%3bcurrID%3aUSD%3bpcFlag%3a0
un.gdp.current.raw <- read_csv(here("Data", "data_raw", "UN data",
"UNdata_Export_20170125_063718499.csv")) %>%
rename(un_code = `Country or Area Code`) %>%
mutate(value.type = "Current")
# https://esa.un.org/unpd/wpp/Download/Standard/Population/
un.pop.raw <- read_excel(here("Data", "data_raw", "UN data",
"WPP2015_POP_F01_1_TOTAL_POPULATION_BOTH_SEXES.xls"),
skip = 16)UN population
un.pop <- un.pop.raw %>%
filter((`Country code` %in% un.codes$un_code)) %>%
select(-c(Index, Variant, Notes, `Major area, region, country or area *`),
un_code = `Country code`) %>%
gather(year, population, -un_code) %>%
left_join(un.codes, by = "un_code") %>% filter(!is.na(iso3)) %>%
mutate(year = as.integer(year),
un.population = population * 1000) %>% # Values are in 1000s
left_join(gw.codes, by = "iso3") %>%
# Get rid of tiny countries
filter(!is.na(cowcode)) %>%
select(cowcode, year, un.population)
un.pop %>% glimpse()## Observations: 12,804
## Variables: 3
## $ cowcode <dbl> 516, 581, 522, 531, 530, 501, 580, 553, 590, 541, 517, 591, 520, 626, 500, 510, 551, 552, 540...
## $ year <int> 1950, 1950, 1950, 1950, 1950, 1950, 1950, 1950, 1950, 1950, 1950, 1950, 1950, 1950, 1950, 195...
## $ un.population <dbl> 2308923, 156334, 62001, 1142150, 18128034, 6076757, 4083554, 2953871, 493254, 6313290, 218618...UN GDP
un.gdp <- bind_rows(un.gdp.raw, un.gdp.current.raw) %>%
filter(Item %in% c("Gross Domestic Product (GDP)",
"Exports of goods and services",
"Imports of goods and services")) %>%
left_join(un.codes, by = "un_code") %>%
# Fix weird edge cases with country names
# Get rid of Former Netherlands Antilles, Zanzibar, Former Czechoslovakia,
# Former Yugoslavia, and Former USSR
filter(!(un_code %in% c(530, 836, 200, 890, 810))) %>%
# Add code for Kosovo, Tanzania, and Sudan
mutate(iso3 = ifelse(un_code == 412, "XKK", iso3),
iso3 = ifelse(un_code == 835, "TZA", iso3),
iso3 = ifelse(un_code == 736, "SDN", iso3))
# Combine the two Yemens, then keep one of the values in the two overlapping years
un.gdp.yemen <- un.gdp %>%
filter(un_code %in% c(720, 886)) %>%
group_by(value.type, Item, Year) %>%
mutate(Value.combined = sum(Value)) %>%
ungroup() %>%
filter(un_code == 720) %>%
mutate(iso3 = "YEM") %>%
select(-Value) %>% select(Value = Value.combined, everything()) %>%
bind_rows(filter(un.gdp, un_code == 887)) %>%
group_by(value.type, Item, Year) %>%
slice(1) %>% ungroup()
# Keep one of the Ethiopias' GDP, since they overlap for a few years
un.gdp.ethiopia <- un.gdp %>%
filter(un_code %in% c(230, 231)) %>%
group_by(value.type, Item, Year) %>%
# mutate(Value.combined = mean(Value)) %>%
slice(1) %>% ungroup() %>%
mutate(iso3 = "ETH")
un.gdp.clean <- un.gdp %>%
# Remove Yemens and Ethiopias
filter(!(un_code %in% c(720, 886, 230, 231))) %>%
# Add consolidated Yemens and Ethiopias
bind_rows(un.gdp.yemen) %>%
bind_rows(un.gdp.ethiopia) %>%
left_join(gw.codes, by = "iso3") %>%
# Get rid of tiny places
filter(!is.na(cowcode))
un.gdp.wide <- un.gdp.clean %>%
select(cowcode, year = Year, Item, Value, value.type) %>%
unite(temp, value.type, Item) %>%
# Temporary id column for spreading correctly
group_by(temp) %>% mutate(id = 1:n()) %>%
spread(temp, Value) %>% select(-id) %>% ungroup() %>%
rename(exports.constant.2005 = `Constant_Exports of goods and services`,
imports.constant.2005 = `Constant_Imports of goods and services`,
gdp.constant.2005 = `Constant_Gross Domestic Product (GDP)`,
exports.current = `Current_Exports of goods and services`,
imports.current = `Current_Imports of goods and services`,
gdp.current = `Current_Gross Domestic Product (GDP)`) %>%
mutate(gdp.deflator = gdp.current / gdp.constant.2005 * 100)
# Rescale the 2005 data to 2011 to match AidData
#
# Deflator = current GDP / constant GDP * 100
# Current GDP in year_t * (deflator in year_target / deflator in year_t)
un.gdp.rescaled <- un.gdp.wide %>%
left_join(select(filter(un.gdp.wide, year == 2011),
cowcode, deflator.target.year = gdp.deflator),
by = "cowcode") %>%
mutate(un.gdp = gdp.current * (deflator.target.year / gdp.deflator),
un.trade.pct.gdp = (imports.current + exports.current) / gdp.current)
un.gdp.final <- un.gdp.rescaled %>%
select(cowcode, year, un.trade.pct.gdp, un.gdp) %>%
group_by(cowcode, year) %>%
slice(1) %>% ungroup()
un.gdp.final %>% head(50) %>% datatable(extensions = "Responsive")## Observations: 8,217
## Variables: 4
## $ cowcode <dbl> 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, ...
## $ year <int> 1970, 1971, 1972, 1973, 1974, 1975, 1976, 1977, 1978, 1979, 1980, 1981, 1982, 1983, 1984, ...
## $ un.trade.pct.gdp <dbl> 0.1073520, 0.1072958, 0.1130692, 0.1305565, 0.1641271, 0.1547753, 0.1600980, 0.1638063, 0....
## $ un.gdp <dbl> 4873912132562, 5034354414646, 5298624445277, 5597607036617, 5568679902693, 5557625605087, ...World Bank indicators
wdi.indicators <- c("NY.GDP.PCAP.KD", # GDP per capita (constant 2010 USD)
"NY.GDP.MKTP.CD", # GDP (current dollars)
"NY.GDP.MKTP.KD", # GDP (constant 2010 USD)
"SP.POP.TOTL", # Population, total
"NE.TRD.GNFS.ZS") # Trade as % of GDP
wdi.path <- here("data", "data_cache", "wdi.rds")
if (!file.exists(wdi.path)) {
# Get all countries and regions because the World Bank chokes on ISO codes like
# XK for Kosovo, even though it returns data for Kosovo with the XK code
# ¯\_(ツ)_/¯
wdi.raw <- WDI(country = "all", wdi.indicators,
extra = FALSE, start = 1980, end = 2015)
saveRDS(wdi.raw, wdi.path)
} else {
wdi.raw <- readRDS(wdi.path)
}
# Filter countries here instead
wdi.clean.intermediate <- wdi.raw %>%
filter(iso2c %in% unique(gw.codes$iso2)) %>%
arrange(iso2c, year) %>%
rename(gdp.capita.2010 = NY.GDP.PCAP.KD,
gdp.2010 = NY.GDP.MKTP.KD, gdp.current = NY.GDP.MKTP.CD,
population = SP.POP.TOTL, trade.pct.gdp = NE.TRD.GNFS.ZS) %>%
mutate(gdp.deflator = gdp.current / gdp.2010 * 100)
# Rescale the 2010 data to 2011 to match AidData
#
# Deflator = current GDP / constant GDP * 100
# Current GDP in year_t * (deflator in year_target / deflator in year_t)
wdi.rescaled <- wdi.clean.intermediate %>%
left_join(select(filter(wdi.clean.intermediate, year == 2011),
iso2c, deflator.target.year = gdp.deflator),
by = "iso2c") %>%
mutate(gdp = gdp.current * (deflator.target.year / gdp.deflator))
wdi.clean <- wdi.rescaled %>%
left_join(select(gw.codes, iso2, cowcode), by = c("iso2c" = "iso2")) %>%
select(-c(gdp.capita.2010, gdp.current, gdp.2010,
gdp.deflator, deflator.target.year))
wdi.clean %>% head(100) %>% datatable(extensions = "Responsive")## Observations: 7,020
## Variables: 7
## $ iso2c <chr> "AD", "AD", "AD", "AD", "AD", "AD", "AD", "AD", "AD", "AD", "AD", "AD", "AD", "AD", "AD", "AD...
## $ country <chr> "Andorra", "Andorra", "Andorra", "Andorra", "Andorra", "Andorra", "Andorra", "Andorra", "Ando...
## $ year <dbl> 1980, 1981, 1982, 1983, 1984, 1985, 1986, 1987, 1988, 1989, 1990, 1991, 1992, 1993, 1994, 199...
## $ population <dbl> 36067, 37500, 39114, 40867, 42706, 44600, 46517, 48455, 50434, 52448, 54509, 56671, 58888, 60...
## $ trade.pct.gdp <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N...
## $ gdp <dbl> 1571153915, 1569072537, 1588630416, 1616751054, 1645604995, 1683806613, 1738586260, 183502776...
## $ cowcode <dbl> 232, 232, 232, 232, 232, 232, 232, 232, 232, 232, 232, 232, 232, 232, 232, 232, 232, 232, 232...Former colonial status
Paul Hensel has collected data on each country’s former colonial status, indicating both a given country’s primary colonial ruler and the last colonial ruler prior to independence. The dataset also includes an estimated date of independence.
Colonial status could matter for aid, with donor countries favoring former colonies (i.e. France might be more likely to donate to former African colonies; Spain might be more likely to donate to Latin America; etc.)
Because there are so many columns, the data below has been horizontally collapsed—click on the green plus sign to see the rest of the data in a row.
col.url <- "http://www.paulhensel.org/Data/colhist.zip"
col.path <- here("Data", "data_raw")
col.zip.name <- basename(col.url)
col.name <- tools::file_path_sans_ext(col.zip.name)
col.final.name <- "coldata100.csv"
col.full.path <- file.path(col.path, "ICOW Colonial History 1.0",
col.final.name)
# Download ICOW data if needed
if (!file.exists(col.full.path)) {
col.get <- GET(col.url,
write_disk(file.path(col.path, col.zip.name),
overwrite = TRUE),
progress())
unzip(file.path(col.path, col.zip.name), exdir = col.path)
file.remove(file.path(col.path, col.zip.name))
}
col.raw <- read_csv(col.full.path, na = "-9",
col_types = cols(
State = col_integer(),
Name = col_character(),
ColRuler = col_integer(),
IndFrom = col_integer(),
IndDate = col_integer(),
IndViol = col_integer(),
IndType = col_integer(),
SecFrom = col_integer(),
SecDate = col_integer(),
SecViol = col_integer(),
Into = col_integer(),
IntoDate = col_integer(),
COWsys = col_integer(),
GWsys = col_integer(),
Notes = col_character()
))
col.clean <- col.raw %>%
# Independence dates with unknown months are coded as 00, which makes
# lubridate choke. So change those to 01
mutate(IndDate.clean = str_replace(IndDate, "00$", "01")) %>%
# Years less than 1000 make lubridate choke, so preface them with a 0
mutate(IndDate.clean = ifelse(nchar(IndDate.clean) == 5,
paste0(0, IndDate.clean), IndDate.clean)) %>%
mutate(IndDate = ymd(paste0(IndDate.clean, "01"))) %>%
left_join(select(gw.codes, iso3, cowcode, country.name),
by = c("State" = "cowcode")) %>%
left_join(select(gw.codes, col.ruler.cowcode = cowcode,
col.ruler.iso3 = iso3,
col.ruler.country.name = country.name),
by = c("ColRuler" = "col.ruler.cowcode")) %>%
left_join(select(gw.codes, ind.from.cowcode = cowcode,
ind.from.iso3 = iso3,
ind.from.country.name = country.name),
by = c("IndFrom" = "ind.from.cowcode")) %>%
select(cowcode = State, country.name, col.ind.date = IndDate,
col.ruler.country.name, col.ruler.cowcode = ColRuler, col.ruler.iso3,
ind.from.country.name, ind.from.cowcode = IndFrom, ind.from.iso3) %>%
# Get rid of smaller ancient states like Tuscany, Parma, and Zanzibar
filter(!is.na(country.name)) %>%
# Add country names and ISO codes for ancient places
mutate(col.ruler.country.name = ifelse(col.ruler.cowcode == 300,
"Austria-Hungary",
col.ruler.country.name),
col.ruler.iso3 = ifelse(col.ruler.cowcode == 300,
"AUH",
col.ruler.country.name)) %>%
mutate(ind.from.country.name = case_when(
.$ind.from.cowcode == 89 ~ "United Provinces of Central America",
.$ind.from.cowcode == 300 ~ "Austria-Hungary",
.$ind.from.cowcode == 315 ~ "Czechoslovakia",
.$ind.from.cowcode == 678 ~ "Yemen (Arab Republic of Yemen)",
.$ind.from.cowcode == 730 ~ "Korea",
is.na(.$ind.from.cowcode) ~ NA_character_,
TRUE ~ .$ind.from.country.name
)) %>%
mutate(ind.from.iso3 = case_when(
.$ind.from.cowcode == 89 ~ "UPC",
.$ind.from.cowcode == 300 ~ "AUH",
.$ind.from.cowcode == 315 ~ "CZE",
.$ind.from.cowcode == 678 ~ "YEM",
.$ind.from.cowcode == 730 ~ "KOR",
is.na(.$ind.from.cowcode) ~ NA_character_,
TRUE ~ .$ind.from.iso3
)) %>%
# Mark never modernly colonzied countries as such instead of missing
mutate(col.ruler.country.name = ifelse(is.na(col.ruler.cowcode),
"Not recently colonized",
col.ruler.country.name),
col.ruler.iso3 = ifelse(is.na(col.ruler.cowcode),
"ZZZ", col.ruler.iso3),
col.ruler.cowcode = ifelse(is.na(col.ruler.cowcode),
999, col.ruler.cowcode),
ind.from.country.name = ifelse(is.na(ind.from.cowcode),
"Already independent",
ind.from.country.name),
ind.from.iso3 = ifelse(is.na(ind.from.cowcode),
"ZZZ", ind.from.iso3),
ind.from.cowcode = ifelse(is.na(ind.from.cowcode),
999, ind.from.cowcode)) %>%
# Serbia isn't included in the colonial ruler data because it's combined with
# Yugoslavia and Montenegro. So here we duplicate the "Croatia" row for
# Serbia to pick up the colonial relationship (even though it's technically
# not 100% correct)
bind_rows(filter(., cowcode == 344) %>%
mutate(cowcode = 340, country.name = "Serbia"))Colonial status data
Summary of clean data
## Observations: 195
## Variables: 9
## $ cowcode <dbl> 2, 20, 31, 40, 41, 42, 51, 52, 53, 54, 55, 56, 57, 58, 60, 70, 80, 90, 91, 92, 93, 9...
## $ country.name <chr> "United States", "Canada", "Bahamas", "Cuba", "Haiti", "Dominican Republic", "Jamaic...
## $ col.ind.date <date> 1783-09-01, 1867-07-01, 1973-07-01, 1902-05-01, 1804-01-01, 1844-02-01, 1962-08-01,...
## $ col.ruler.country.name <chr> "United Kingdom", "United Kingdom", "United Kingdom", "Spain", "France", "Spain", "U...
## $ col.ruler.cowcode <dbl> 200, 200, 200, 230, 220, 230, 200, 200, 200, 200, 200, 200, 200, 200, 200, 230, 200,...
## $ col.ruler.iso3 <chr> "United Kingdom", "United Kingdom", "United Kingdom", "Spain", "France", "Spain", "U...
## $ ind.from.country.name <chr> "United Kingdom", "United Kingdom", "United Kingdom", "United States", "France", "Ha...
## $ ind.from.cowcode <dbl> 200, 200, 200, 2, 220, 41, 200, 200, 200, 200, 200, 200, 200, 200, 200, 230, 200, 89...
## $ ind.from.iso3 <chr> "GBR", "GBR", "GBR", "USA", "FRA", "HTI", "GBR", "GBR", "GBR", "GBR", "GBR", "GBR", ...Conflict
The UCDP/PRIO Armed Conflict Dataset tracks a ton of conflict-releated data, including reasons for the conflict, parties in the conflict, intensity of the conflict, and deaths in the conflict. We’re only interested in whether a conflict happened in a given year (or in the past 5 years), so here we simply create an indicator variable for whether there was internal conflict in a country-year (conflict type = 3).
# Uppsala conflict data
# http://www.pcr.uu.se/research/ucdp/datasets/generate_your_own_datasets/dynamic_datasets/
# conflicts <- read_tsv(here("Data", "data_raw", "External",
# "Uppsala", "ywd_dataset.csv")) %>%
# mutate(intensity = factor(intensity,
# levels = c("No", "Minor", "Intermediate", "War"),
# ordered = TRUE))
conflicts.url <- "http://ucdp.uu.se/downloads/ucdpprio/ucdp-prio-acd-4-2016.csv"
conflicts.path <- here("Data", "data_raw", "UCDP")
if (!file.exists(file.path(conflicts.path, basename(conflicts.url)))) {
GET(conflicts.url,
write_disk(file.path(conflicts.path, basename(conflicts.url)),
overwrite = TRUE))
}
conflicts.raw <- read_csv(file.path(conflicts.path, basename(conflicts.url)))
conflicts <- conflicts.raw %>%
filter(TypeOfConflict == 3) %>%
mutate(gwcode = as.integer(GWNoA)) %>%
group_by(gwcode, Year) %>%
summarise(conflict.internal = n() > 0) %>%
ungroup() %>%
left_join(gw.codes, by = "gwcode") %>%
filter(!is.na(cowcode))
glimpse(conflicts)## Observations: 1,203
## Variables: 7
## $ gwcode <dbl> 40, 40, 40, 40, 41, 41, 41, 42, 52, 70, 70, 90, 90, 90, 90, 90, 90, 90, 90, 90, 90, 90, 9...
## $ Year <int> 1953, 1956, 1957, 1958, 1989, 1991, 2004, 1965, 1990, 1994, 1996, 1949, 1954, 1963, 1965,...
## $ conflict.internal <lgl> TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE,...
## $ country.name <chr> "Cuba", "Cuba", "Cuba", "Cuba", "Haiti", "Haiti", "Haiti", "Dominican Republic", "Trinida...
## $ iso3 <chr> "CUB", "CUB", "CUB", "CUB", "HTI", "HTI", "HTI", "DOM", "TTO", "MEX", "MEX", "GTM", "GTM"...
## $ iso2 <chr> "CU", "CU", "CU", "CU", "HT", "HT", "HT", "DO", "TT", "MX", "MX", "GT", "GT", "GT", "GT",...
## $ cowcode <dbl> 40, 40, 40, 40, 41, 41, 41, 42, 52, 70, 70, 90, 90, 90, 90, 90, 90, 90, 90, 90, 90, 90, 9...Natural disasters
Natural disaster data comes from the International Disaster Database (EM-DAT). The data includes the number of deaths, injuries, homeless displacements, and monetary losses (in 2000 dollars) for a huge number of natural and technological disasters (see EM-DAT’s full classification).
Natural disasters could matter for aid too, since donor countries might increase their aid to countries suffering more.
EM-DAT does not provide a single link to download their data. Instead, you have to create a query using their advanced search form. We downloaded data using the following query:
- Select all countries from up to 2016
- Select all three disaster classification groups (natural, technological, complex)
- Group results by country name, year, and disaster type
- Download CSV and save in
Data/data_raw/Disasters/Data.csv
# Disaster classification
disaster.classification.raw <- "https://web.archive.org/web/20161203165345/http://www.emdat.be/classification" %>%
read_html() %>%
html_nodes(., xpath = '//*[@id="article-14627"]/div/div/div/div/table[1]') %>%
html_table(header = TRUE)
# For whatever reason, the downloaded data treats technological subgroups as
# main types (e.g. "industrial accident" is listed as a disaster type, not
# "chemical spill" or "fire" or whatever). So, make a new column of the actual
# disaster type using the real type for natural disasters and the subgroup for
# non-natural disasters.
#
# Also, add a couple rows for complex and miscellaneous disasters
disaster.classification <- disaster.classification.raw[[1]] %>%
select(-Definition) %>%
mutate_all(funs(str_trim(str_to_lower(.)))) %>%
# Fix typo
mutate(`Disaster Subgroup` = ifelse(`Disaster Subgroup` == "miscelleanous accident",
"miscellaneous accident", `Disaster Subgroup`)) %>%
mutate(`Actual Type` = ifelse(`Disaster Group` == "natural",
`Disaster Main Type`, `Disaster Subgroup`)) %>%
distinct(`Disaster Group`, `Disaster Subgroup`, `Actual Type`) %>%
bind_rows(tribble(
~`Disaster Group`, ~`Disaster Subgroup`, ~`Actual Type`,
"complex", "complex disasters", "complex disasters"
))
# Disaster data
# http://www.emdat.be/advanced_search/index.html
# Select all countries from whenever to 2016
# Select all three disaster classification groups (natural, technological, complex)
# Group results by country name, year, and disaster type
# Download CSV and save in Data/data_raw/Disasters
disasters.raw <- read_csv(here("Data", "data_raw", "Disasters", "data.csv"),
skip = 1) # First row is junk
# Read in disaster data and join with classification data
disasters <- disasters.raw %>%
mutate(`disaster type` = str_trim(str_to_lower(`disaster type`))) %>%
mutate(`disaster type` = ifelse(`disaster type` == "mass movement (dry)",
"mass movement", `disaster type`)) %>%
left_join(disaster.classification, by = c("disaster type" = "Actual Type")) %>%
# Get rid of tiny countries
filter(!(iso %in% c("AIA", "ANT", "ASM", "AZO", "BMU", "COK", "CSK", "CYM",
"DDR", "GLP", "GUF", "GUM", "MAC", "MNP", "MSR", "MTQ",
"MYT", "NCL", "NIU", "PRI", "PYF", "REU", "SHN", "SPI",
"TCA", "TKL", "VGB", "VIR", "WLF", "YMD"))) %>%
mutate(iso = str_to_upper(iso),
cowcode = countrycode(iso, "iso3c", "cown",
custom_match = c(HKG = 715, DFR = 255, PSE = 669,
SUN = 365, YMN = 679, SRB = 340,
SCG = 345, YUG = 345))) %>%
# Fix missing names (collapsing old Germanies, Yemens, and USSR)
mutate(country_name = countrycode(iso, "iso3c", "country.name",
custom_match = c(DFR = "Germany",
SCG = "Yugoslavia",
YUG = "Yugoslavia",
SUN = "Russia",
YMN = "Yemen"))) %>%
# Fix ISOs
mutate(iso = countrycode(country_name, "country.name", "iso3c",
custom_match = c(Yugoslavia = "YUG"))) %>%
select(country_name, year, iso3 = iso, cowcode, disaster.type = `disaster type`,
disaster.group = `Disaster Group`, disaster.subgroup = `Disaster Subgroup`,
disaster.occurrence = occurrence, disaster.deaths = `Total deaths`,
disaster.injured = `Injured`, disaster.affected = `Affected`,
disaster.homeless = `Homeless`, disaster.total.affected = `Total affected`,
disaster.total.damage = `Total damage`) %>%
filter(disaster.group != "complex")Clean disaster data
# Summarize disaster variables by country, year, and type/group/subgroup
disaster.vars <- vars(disaster.occurrence, disaster.deaths, disaster.injured,
disaster.affected, disaster.homeless, disaster.total.affected,
disaster.total.damage)
disasters.groups <- disasters %>%
group_by(cowcode, year, disaster.group) %>%
summarise_at(disaster.vars, funs(sum(., na.rm = TRUE))) %>%
ungroup() %>%
gather(key, value, disaster.occurrence, disaster.deaths, disaster.injured,
disaster.affected, disaster.homeless, disaster.total.affected,
disaster.total.damage) %>%
unite(key, disaster.group, key) %>%
spread(key, value) %>%
filter(year > 1980, year < 2016)
disasters.groups %>% head(100) %>% datatable(extensions = "Responsive")Summary of clean data
## Observations: 4,086
## Variables: 16
## $ cowcode <dbl> 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, ...
## $ year <int> 1981, 1982, 1983, 1984, 1985, 1986, 1987, 1988, 1989, 1990, 1991, 199...
## $ natural_disaster.affected <int> 0, 36000, 1500, 14000, 1005550, 2150, 5400, 0, 300, 14188, 2900, 4545...
## $ natural_disaster.deaths <int> 60, 514, 332, 1145, 375, 62, 239, 27, 259, 214, 132, 96, 542, 275, 87...
## $ natural_disaster.homeless <int> 0, 15500, 6000, 5500, 20000, 0, 0, 0, 25015, 0, 300, 250540, 300, 210...
## $ natural_disaster.injured <int> 0, 0, 53, 657, 500, 109, 94, 0, 3857, 30, 200, 481, 430, 7522, 160, 1...
## $ natural_disaster.occurrence <int> 4, 13, 12, 14, 15, 7, 11, 22, 19, 24, 36, 32, 30, 16, 18, 15, 33, 32,...
## $ natural_disaster.total.affected <int> 0, 51500, 7553, 20157, 1026050, 2259, 5494, 0, 29172, 14218, 3400, 29...
## $ natural_disaster.total.damage <int> 861000, 2145900, 5107250, 2287600, 5094100, 1845720, 421000, 0, 13480...
## $ technological_disaster.affected <int> 0, 23000, 2000, 0, 0, 0, 24000, 0, 0, 185, 16000, 0, 0, 9230, 200, 12...
## $ technological_disaster.deaths <int> 26, 153, 0, 0, 12, 138, 296, 58, 344, 216, 170, 59, 172, 271, 41, 404...
## $ technological_disaster.homeless <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 300, 0, 0, 0, 0, 0, 120, 0, 20...
## $ technological_disaster.injured <int> 240, 355, 43, 396, 265, 0, 6, 0, 185, 336, 251, 24, 3156, 649, 485, 4...
## $ technological_disaster.occurrence <int> 3, 5, 1, 3, 3, 3, 10, 4, 16, 8, 10, 3, 7, 13, 7, 10, 6, 9, 12, 7, 9, ...
## $ technological_disaster.total.affected <int> 240, 23355, 2043, 396, 265, 0, 24006, 0, 185, 521, 16251, 24, 3156, 1...
## $ technological_disaster.total.damage <int> 0, 0, 0, 0, 0, 0, 0, 0, 1100000, 220000, 14500, 0, 0, 33000, 0, 35100...Countries to exclude
We exclude long-term consolidated democracies from our analysis, following FinkelPerez-LinanSeligson:2007, 414. These are classified by the World Bank as high income; they score below 3 on Freedom House’s Scale, receive no aid from USAID, and are not newly independent states.
consolidated.democracies <-
data_frame(country.name = c("Andorra", "Australia", "Austria", "Bahamas",
"Barbados", "Belgium", "Canada", "Denmark", "Finland",
"France", "Germany", "Greece", "Grenada", "Iceland",
"Ireland", "Italy", "Japan", "Liechtenstein", "Luxembourg",
"Malta", "Monaco", "Netherlands", "New Zealand", "Norway",
"San Marino", "Spain", "Sweden", "Switzerland",
"United Kingdom", "United States of America")) %>%
mutate(iso3 = countrycode(country.name, "country.name", "iso3c"),
cowcode = countrycode(country.name, "country.name", "cown"))
consolidated.democracies %>% pandoc.table()| country.name | iso3 | cowcode |
|---|---|---|
| Andorra | AND | 232 |
| Australia | AUS | 900 |
| Austria | AUT | 305 |
| Bahamas | BHS | 31 |
| Barbados | BRB | 53 |
| Belgium | BEL | 211 |
| Canada | CAN | 20 |
| Denmark | DNK | 390 |
| Finland | FIN | 375 |
| France | FRA | 220 |
| Germany | DEU | 255 |
| Greece | GRC | 350 |
| Grenada | GRD | 55 |
| Iceland | ISL | 395 |
| Ireland | IRL | 205 |
| Italy | ITA | 325 |
| Japan | JPN | 740 |
| Liechtenstein | LIE | 223 |
| Luxembourg | LUX | 212 |
| Malta | MLT | 338 |
| Monaco | MCO | 221 |
| Netherlands | NLD | 210 |
| New Zealand | NZL | 920 |
| Norway | NOR | 385 |
| San Marino | SMR | 331 |
| Spain | ESP | 230 |
| Sweden | SWE | 380 |
| Switzerland | CHE | 225 |
| United Kingdom | GBR | 200 |
| United States of America | USA | 2 |
Final clean combined data
With both donor- and country-level data, we have lots of different options for analysis. Since our hypotheses deal with questions of donor responses, the data we use to model donor responses uses donor-years as the unit of observation. Not all donors give money to the same countries, so this final data is not a complete panel (i.e. it does not include every combination of donors and years), which will pose some interesting methodological issues when modeling.
In H3 we hypothesize that more aid will be allocated to international or US-based NGOs than domestic NGOs in response to harsher anti-NGO restrictions. While AidData unfortunately does not categorize aid by channel (i.e. aid given to international vs. US vs. domestic NGOs), USAID does. For this hypothesis, then we only look at aid given by USAID, not the rest of the OECD. As with the proportion of contentious aid, we create similar variables to measure the proporition of aid given to international NGOs, US-based NGOs, and both international and US-based NGOs.
# Donor data
donor.level.data.raw <- aiddata.final %>%
filter(cowcode %in% dcjw.full$cowcode) %>%
filter(year > 1980) %>%
filter(oda > 0) %>% # Only look at positive aid
mutate(oda_log = log1p(oda))
# Create fake country codes for non-country donors
fake.codes <- donor.level.data.raw %>%
distinct(donor, donor.type) %>%
filter(donor.type != "Country") %>%
arrange(donor.type) %>% select(-donor.type) %>%
mutate(fake.donor.cowcode = 2001:(2000 + n()),
fake.donor.iso3 = paste0("Z", str_sub(fake.donor.cowcode, 3)))
donor.level.data <- donor.level.data.raw %>%
left_join(fake.codes, by = "donor") %>%
mutate(donor.cowcode = ifelse(is.na(donor.cowcode),
fake.donor.cowcode,
donor.cowcode),
donor.iso3 = ifelse(is.na(donor.iso3),
fake.donor.iso3,
donor.iso3)) %>%
select(-starts_with("fake"))
# USAID data
# USAID's conversion to constant 2015 dollars doesn't seem to take country differences into account—the deflator for each country in 2011 is essentially 96.65. When there are differences, it's because of floating point issues (like, if there are tiny grants of $3, there aren't enough decimal points to get the fraction to 96.65). So we just take the median value of the deflator for all countries and all grants and use that.
#
# Rescale the 2015 data to 2011 to match AidData
#
# Deflator = current aid / constant aid * 100
# Current aid in year_t * (deflator in year_target / deflator in year_t)
usaid.deflator.2011 <- usaid.clean %>%
filter(year == 2011) %>%
summarise(deflator.target.year = median(aid.deflator, na.rm = TRUE)) %>%
as.numeric()
donor.level.data.usaid <- usaid.clean %>%
filter(cowcode %in% dcjw.full$cowcode) %>%
filter(year > 1980) %>%
filter(oda.us.current > 0) %>%
mutate(oda.us.2011 = oda.us.current * (usaid.deflator.2011 / aid.deflator))
usaid.by.country.total <- donor.level.data.usaid %>%
group_by(cowcode, year) %>%
summarise(oda.us = sum(oda.us.2011, na.rm = TRUE))
usaid.by.country.channel <- donor.level.data.usaid %>%
gather(key, value, c(channel.ngo.us, channel.ngo.int, channel.ngo.dom)) %>%
group_by(cowcode, year, key, value) %>%
summarise(total.oda.us = sum(oda.us.2011, na.rm = TRUE)) %>%
ungroup() %>%
unite(channel, key, value) %>%
filter(str_detect(channel, "TRUE")) %>%
mutate(channel = str_replace(channel, "channel", "oda.us"),
channel = str_replace(channel, "_TRUE", "")) %>%
spread(channel, total.oda.us, fill = 0)
# Country data
# Use DCJW 1980+ as base data, since we're limited to the countries included there
country.level.data <- dcjw.full %>%
mutate(ever.dac.eligible = cowcode %in% ever.dac.eligible) %>%
filter(!(cowcode %in% consolidated.democracies$cowcode)) %>%
left_join(select(vdem.cso, year, cowcode, csre,
polity = e_fh_ipolity2,
corruption = v2x_corr,
govt.effectiveness = e_wbgi_gee),
by = c("cowcode", "year")) %>%
mutate(corruption = corruption * 10) %>%
left_join(select(wdi.clean, -c(iso2c, country)), by = c("cowcode", "year")) %>%
# Bring in UN population data
left_join(un.pop, by = c("cowcode", "year")) %>%
# Prefer UN population data, but use World Bank if missing
mutate(population = ifelse(is.na(un.population), population, un.population)) %>%
# Bring in UN GDP data
# Prefer UN GDP data, but use World Bank if missing
left_join(un.gdp.final, by = c("cowcode", "year")) %>%
mutate(un.trade.pct.gdp = un.trade.pct.gdp * 100) %>%
mutate(gdp = ifelse(is.na(un.gdp), gdp, un.gdp),
trade.pct.gdp = ifelse(is.na(un.trade.pct.gdp),
trade.pct.gdp, un.trade.pct.gdp)) %>%
select(-starts_with("un.")) %>%
left_join(select(col.clean, -country.name), by = c("cowcode")) %>%
left_join(select(disasters.groups, cowcode, year, starts_with("natural_")),
by = c("cowcode", "year")) %>%
# NAs in disasters are really 0, especially when occurrence is 0
mutate_at(vars(starts_with("natural_")), funs(ifelse(is.na(.), 0, .))) %>%
# Treat NAs in conflicts as FALSE
left_join(select(conflicts, cowcode, year = Year, conflict.internal),
by = c("cowcode", "year")) %>%
mutate(conflict.internal = ifelse(is.na(conflict.internal),
FALSE, conflict.internal)) %>%
# Add indicator for post-Cold War, since all the former Soviet republics have
# no GDP data before 1990
mutate(post.1989 = year >= 1990)
testthat::expect_equal(nrow(country.level.data),
nrow(filter(dcjw.full,
!(cowcode %in% consolidated.democracies$cowcode))))
# Combine country and donor data
donor.country.data <- donor.level.data %>%
left_join(select(country.level.data, -country.name),
by = c("year", "cowcode")) %>%
mutate(former.colony = donor.cowcode == col.ruler.cowcode |
donor.cowcode == ind.from.cowcode) %>%
arrange(donor, year)
testthat::expect_equal(nrow(donor.country.data), nrow(donor.level.data))
# Calculate different versions of aid variables
aid.by.country.total <- donor.country.data %>%
group_by(cowcode, year) %>%
summarise(total.oda = sum(oda, na.rm = TRUE))
aid.by.country.purpose <- donor.country.data %>%
group_by(cowcode, year, purpose.contentiousness) %>%
summarise(total.oda = sum(oda, na.rm = TRUE)) %>%
spread(purpose.contentiousness, total.oda, fill = 0) %>%
rename(oda.contentious.high = High,
oda.contentious.low = Low)
# Neighbor aid
# Join total oda to the huge distances dataframe and calculate total and
# average aid to neighboring countries
neighbor.oda <- all.distances %>%
filter(is.rough.neighbor) %>%
left_join(aid.by.country.total, by = c("cowcode.other" = "cowcode",
"year")) %>%
filter(cowcode.self != cowcode.other) %>%
group_by(year, cowcode.self) %>%
summarise_at(vars(total.oda),
funs(nb_oda.sum = sum(., na.rm = TRUE),
nb_oda.avg = mean(., na.rm = TRUE),
nb_oda.sd = sd(., na.rm = TRUE),
nb_oda.n = n())) %>%
rename(cowcode = cowcode.self) %>%
filter(year < 2014) %>%
mutate_at(vars(starts_with("nb")),
funs(ifelse(is.nan(.), 0, .)))Missingness and imputation
The donor data is complete with no missing variables(!).
The country-level panel data is relatively complete, with only a few variables suffering from missing data, mostly from the World Bank and V-Dem.
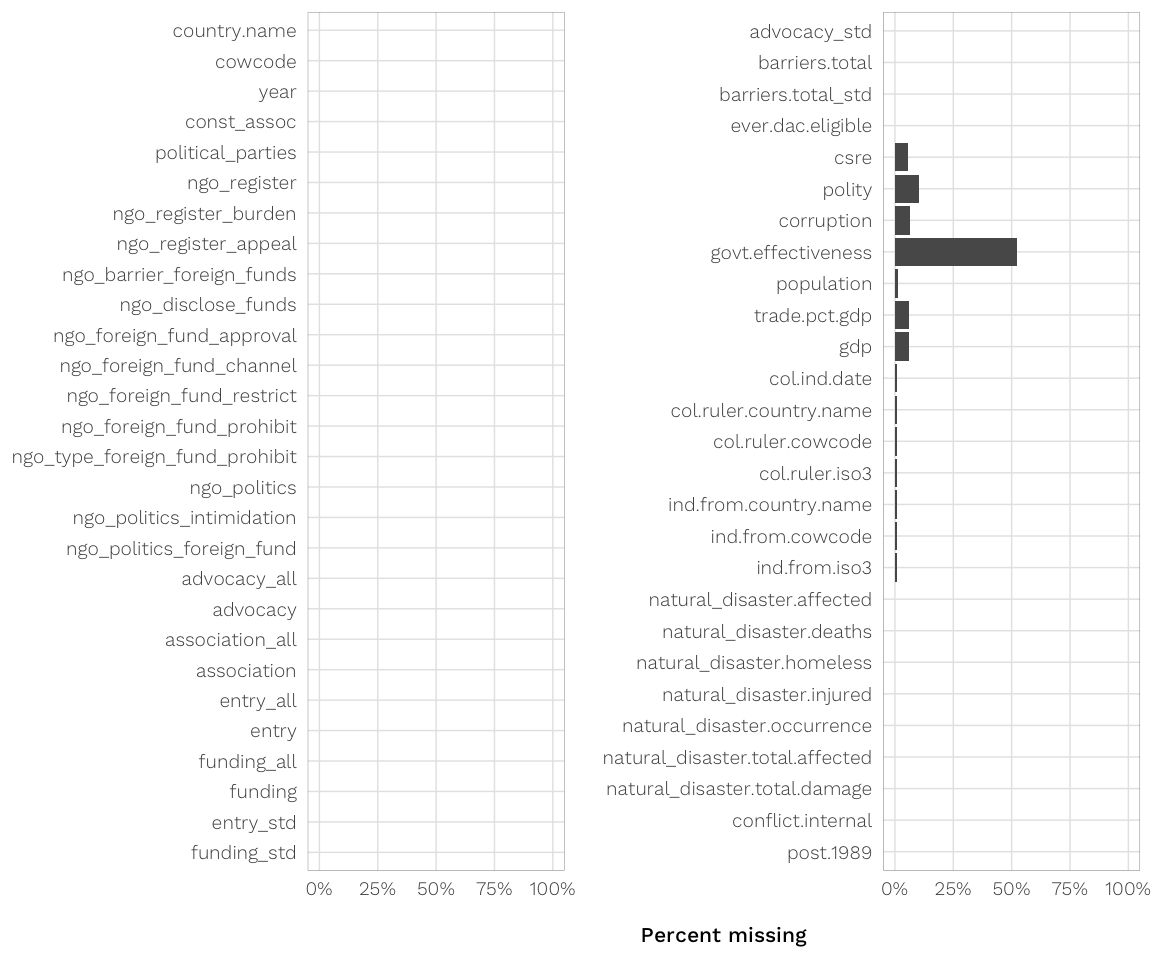
That said, it’s still helpful to impute the missing variables, especially since polity, corruption, and GDP are all main controls (plus Lall 2016 shows the downsides of not imputing). So, we use Amelia to impute missing variables.
We ultimately create a long data frame with each of the imputed datasets stacked under the original dataset. Datasets are identified with the column m, like so:
stacked.data <- tribble(
~m, ~country, ~year, ~variable,
"original", "Afghanistan", "2000", 5,
"original", "Afghanistan", "2001", NA,
"original", "Afghanistan", "2002", 6,
"...", "...", "...", NA,
"imp1", "Afghanistan", "2000", 5,
"imp1", "Afghanistan", "2001", 5.3,
"imp1", "Afghanistan", "2002", 6,
"...", "...", "...", NA,
"imp2", "Afghanistan", "2000", 5,
"imp2", "Afghanistan", "2001", 5.8,
"imp2", "Afghanistan", "2002", 6,
"...", "...", "...", NA
)
pandoc.table(stacked.data)| m | country | year | variable |
|---|---|---|---|
| original | Afghanistan | 2000 | 5 |
| original | Afghanistan | 2001 | NA |
| original | Afghanistan | 2002 | 6 |
| … | … | … | NA |
| imp1 | Afghanistan | 2000 | 5 |
| imp1 | Afghanistan | 2001 | 5.3 |
| imp1 | Afghanistan | 2002 | 6 |
| … | … | … | NA |
| imp2 | Afghanistan | 2000 | 5 |
| imp2 | Afghanistan | 2001 | 5.8 |
| imp2 | Afghanistan | 2002 | 6 |
| … | … | … | NA |
We estimate models for each of the imputed datasets and recombine their results. We use amelia::mi.meld() to combine estimates and standard errors from frequentist models (following King et al. 2001), and we merge the MCMC chains from Bayesian models (see also this).
We varied the number of datasets we imputed (m = 5 and m = 10) and found only trivial differences in coefficients between 5 and 10 datasets (as predicted by King et al.). In the interest of computational efficiency and speed, we only use 5 imputed datasets. To avoid needing to recreate both 5 and 10 datasets, we impute 10, but only use the first 5 for analysis.
imputation.rds <- here("Data", "data_cache", "country_aid_data_imputed_long.rds")
if (file.exists(imputation.rds)) {
country.aid.data.imputed.long <- readRDS(here("Data", "data_cache",
"country_aid_data_imputed_long.rds"))
} else {
# THIS TAKES SO LONG.
#
# Combine aid data with country data
country.aid.data.pre.transformation <- country.level.data %>%
filter(year < 2014) %>% # There's no aid data after 2013
left_join(aid.by.country.total, by = c("cowcode", "year")) %>%
left_join(aid.by.country.purpose, by = c("cowcode", "year")) %>%
left_join(neighbor.oda, by = c("cowcode", "year")) %>%
left_join(usaid.by.country.total, by = c("cowcode", "year")) %>%
left_join(usaid.by.country.channel, by = c("cowcode", "year")) %>%
mutate_at(vars(contains("oda")), funs(ifelse(is.na(.), 0, .)))
# Missing Polity values are easy—most are just one-year gaps with the same
# value before and after, so interpolate the missing values
polity.imputed <- country.aid.data.pre.transformation %>%
select(cowcode, year, polity) %>%
group_by(cowcode) %>%
mutate(impute.polity = !all(is.na(polity))) %>%
mutate(polity = ifelse(impute.polity, na.interpolation(polity), polity)) %>%
ungroup() %>% select(-impute.polity)
country.aid.data.pre.amelia <- country.aid.data.pre.transformation %>%
select(-polity) %>%
left_join(polity.imputed, by = c("cowcode", "year"))
# Select the variables to not include in the imputation model. Left join this
# data frame later with the imputed data
vars.to.include.in.imputation <- country.aid.data.pre.amelia %>%
select(csre, corruption, govt.effectiveness, population, trade.pct.gdp,
gdp, starts_with("natural"), total.oda, polity) %>%
select(-natural_disaster.total.affected)
# Vector of variables to include
vars.to.include.sans.ids <- vars.to.include.in.imputation %>% colnames()
# Stripped down dataset for imputation
country.aid.data.to.impute <- country.aid.data.pre.amelia %>%
select(year, cowcode, one_of(vars.to.include.sans.ids))
# Imputation model tweaks
# Variables that will be lagged and leaded
vars.to.lag.lead <- c("csre", "polity", "corruption", "population",
"trade.pct.gdp", "gdp")
# Variables that will be logged
vars.to.log <- c("gdp", "population")
# Run imputation model
set.seed(my.seed)
country.aid.data.amelia <- amelia(country.aid.data.to.impute,
m = 10, ts = "year", cs = "cowcode",
intercs = TRUE,
empri = 0.03 * nrow(country.aid.data.to.impute),
p2s = 1,
lags = vars.to.lag.lead,
leads = vars.to.lag.lead,
logs = vars.to.log,
parallel = "multicore",
ncpus = parallel::detectCores())
# Append list of imputed datasets to original in a long data frame and bring
# back the omitted variables
country.aid.data.imputed.long <-
bind_rows(mutate(country.aid.data.to.impute, m = "original"),
bind_rows(unclass(country.aid.data.amelia$imputations), .id = "m")) %>%
select(m, everything()) %>%
left_join(select(country.aid.data.pre.amelia,
-one_of(vars.to.include.sans.ids)), by = c("cowcode", "year"))
saveRDS(country.aid.data.imputed.long, here("Data", "data_cache",
"country_aid_data_imputed_long.rds"))
}# Log, lead, and lag everything based on the big dataset of imputations
# pct.change <- function(x) {
# (lead(x) / x) - 1
# }
# Determine if any of the values in the last k rows are TRUE
check_last_k <- function(x, k) {
# This creates a matrix with a column for each lag value (e.g. column 1 = lag
# 0, column 2 = lag 1, etc.)
all.lags <- sapply(0:k, FUN = function(k) lag(x, k))
# Marke TRUE if any of the columns have TRUE in them
any.true.in.window <- apply(all.lags, MARGIN = 1, FUN = any, na.rm = TRUE)
return(any.true.in.window)
}
country.aid.data <- country.aid.data.imputed.long %>%
mutate(gdp.capita = gdp / population) %>%
mutate_at(vars(gdp.capita, gdp, population), funs(log = log1p)) %>%
# Calculate prop.contentious here since there should be some missing values
# (since there are country-years where there's no ODA)
mutate(prop.contentious = oda.contentious.high /
(oda.contentious.low + oda.contentious.high)) %>%
mutate(prop.contentious = ifelse(is.nan(prop.contentious),
NA, prop.contentious)) %>%
mutate(prop.contentious_logit = car::logit(prop.contentious, adjust = .001)) %>%
# Calculate prop.ngo* here since there should also be some missing values
mutate(prop.ngo.int = oda.us.ngo.int / oda.us,
prop.ngo.us = oda.us.ngo.us / oda.us,
prop.ngo.dom = oda.us.ngo.dom / oda.us,
prop.ngo.foreign = (oda.us.ngo.int + oda.us.ngo.us) / oda.us) %>%
# prop.ngo.dom.foreign = oda.us.ngo.dom / (oda.us.ngo.int + oda.us.ngo.us)) %>%
mutate_at(vars(starts_with("prop.ngo")),
funs(ifelse(is.nan(.), NA, .))) %>%
mutate_at(vars(starts_with("prop.ngo")),
funs(logit = car::logit(., adjust = .001))) %>%
mutate_at(vars(total.oda, oda.contentious.high, oda.contentious.low, oda.us,
nb_oda.sum, nb_oda.avg, nb_oda.sd),
funs(log = log1p(.))) %>%
mutate(aid.pct.gdp = total.oda / gdp) %>%
# Lag/lead/diff everything within countries and samples
group_by(m, cowcode) %>%
# Get leaded aid variables
mutate_at(vars(contains("oda"), starts_with("prop.")),
funs(next_year = lead(.), # y_{i, t+1}
after_2 = lead(., 2),
after_5 = lead(., 5))) %>%
# Get diffed leaded aid variables
mutate_at(vars(contains("next_year"), -contains("log")),
funs(diff = lead(.) - ., # (y_{it} - y_{i, t-1})_{t+1}
diff_log = sign(lead(.) - .) *
log1p(abs(lead(.) - .)))) %>% # sign(x) * log1p(abs(x))
mutate_at(vars(contains("after_2"), -contains("log")),
funs(diff = lead(., 2) - .,
diff_log = sign(lead(., 2) - .) *
log1p(abs(lead(., 2) - .)))) %>%
mutate_at(vars(contains("after_5"), -contains("log")),
funs(diff = lead(., 5) - .,
diff_log = sign(lead(., 5) - .) *
log1p(abs(lead(., 2) - .)))) %>%
# Indicate changes in laws
mutate_at(vars(advocacy, association, entry, funding, barriers.total),
funs(new = . - lag(.),
worse = (. - lag(.)) > 0,
cat = cut(. - lag(.),
breaks = c(-Inf, -1, 0, Inf),
labels = c("New better law", "No new laws",
"New worse law"),
ordered_result = TRUE))) %>%
# Determine if there was conflict in the past 5 years
mutate(internal.conflict.past.5 = check_last_k(conflict.internal, 5)) %>%
ungroup()
# Save all 10 imputed datasets
country.aid.data.m10 <- country.aid.data
# Use just the first 5 imputed datasets
country.aid.data <- country.aid.data %>%
filter(!(m %in% paste0("imp", 6:10)))The final imputed dataset still has missing data, but that’s because of (1) prop.contentious, which is missing if there’s no aid in a country year, and (2) lags and leads create missing values at the beginnings and ends of the country windows.
Save final data
Final, clean country- and donor-level datasets are available in Data/data_clean/.