Preliminary analysis
Steven L. Peck and Andrew Heiss
Last run: April 23, 2020
library(tidyverse)
library(brms)
library(modelr)
library(broom)
library(huxtable)
library(scales)
library(here)
CHAINS <- 4
ITER <- 4000
WARMUP <- 2000
BAYES_SEED <- 1234
options(mc.cores = parallel::detectCores())
constraints <- read_rds(here("data", "derived_data", "constraints.rds"))
constraint_levels <- constraints %>%
mutate(constraint_clean = fct_inorder(constraint_clean)) %>%
mutate(levels = map(levels_clean, names)) %>%
unnest(levels) %>%
mutate(levels = fct_inorder(levels))
# TODO: Switch back to full BLS.rds and BHL.rds
BHL <- read_rds(here("data", "derived_data", "BHL_small.rds"))
bhl_long <- BHL %>%
gather(constraint, value, one_of(constraints$constraint)) %>%
left_join(constraints, by = "constraint") %>%
mutate(value = fct_relevel(value, levels(constraint_levels$levels)),
constraint_clean = fct_relevel(constraint_clean,
levels(constraint_levels$constraint_clean)))
BLS <- read_rds(here("data", "derived_data", "BLS_small.rds"))
bls_long <- BLS %>%
gather(constraint, value, one_of(constraints$constraint)) %>%
left_join(constraints, by = "constraint") %>%
mutate(value = fct_relevel(value, levels(constraint_levels$levels)),
constraint_clean = fct_relevel(constraint_clean,
levels(constraint_levels$constraint_clean)))Single conditions, no nested interactions
Landscape fitness
High-low
ggplot(bhl_long, aes(x = value, y = avg_evenness_t, fill = repp)) +
geom_violin(size = 0.25, position = position_dodge(width = 0.75)) +
stat_summary(geom = "point", fun = "mean",
aes(group = repp), position = position_dodge(width = 0.75),
size = 2, pch = 21, fill = "black", color = "white") +
# scale_y_reverse(breaks = seq(0, 0.4, 0.2)) +
# coord_cartesian(ylim = c(0, 0.5)) +
scale_fill_viridis_d(option = "plasma", begin = 0.1, end = 0.9) +
labs(x = NULL, y = "Landscape fitness, linked", fill = NULL,
caption = "Point = mean value") +
facet_wrap(~ constraint_clean, scales = "free_x") +
theme_bw() +
theme(legend.position = "top",
legend.key.size = unit(0.65, "lines"))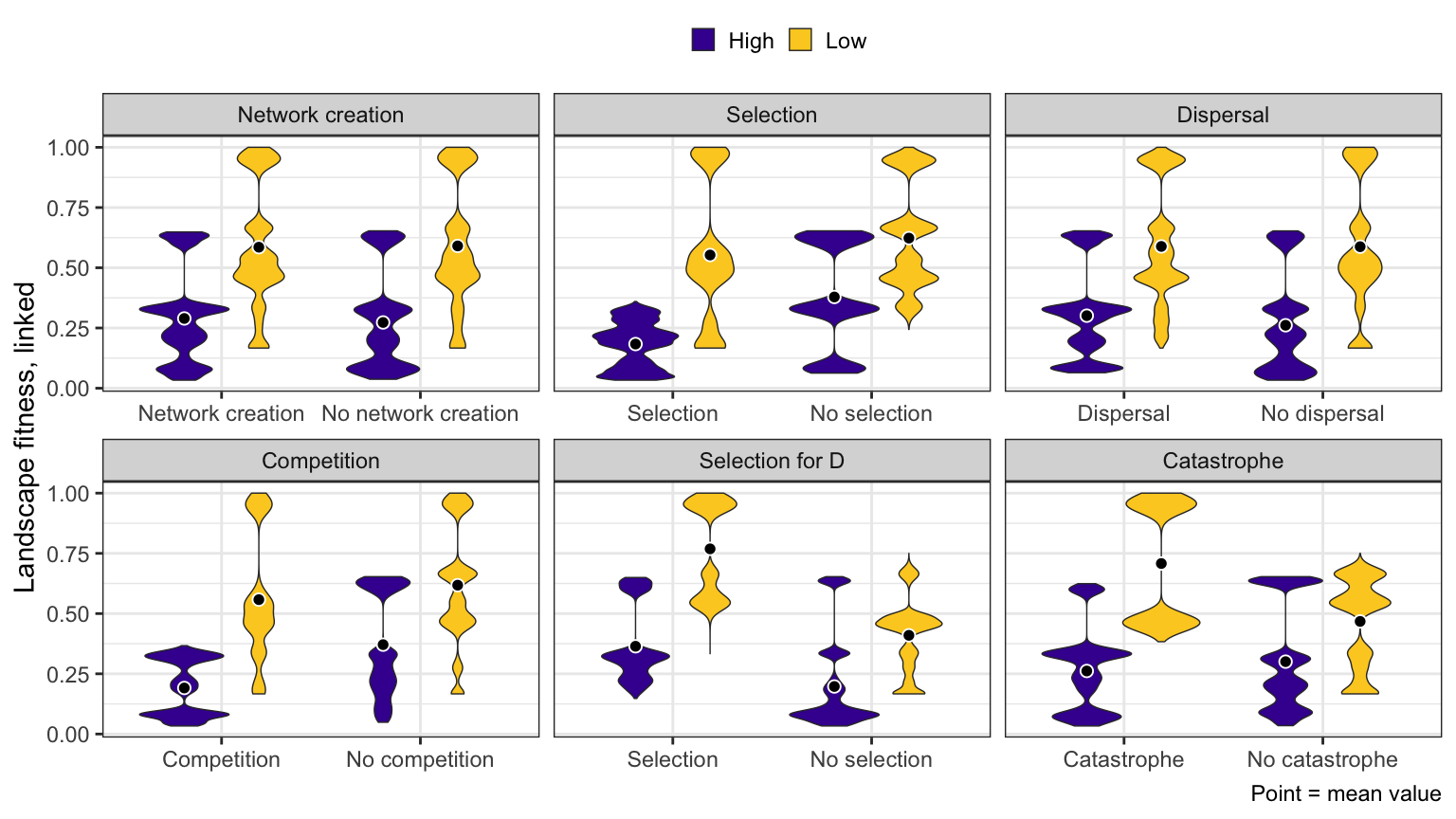
Latin squares
ggplot(bls_long,
aes(x = value, y = landscape_fitness_linked, fill = constraint_clean)) +
geom_violin(size = 0.25) +
stat_summary(geom = "point", fun = "mean",
size = 2, pch = 21, fill = "black", color = "white") +
scale_y_reverse(breaks = seq(0, 0.4, 0.2)) +
coord_cartesian(ylim = c(0, 0.5)) +
scale_fill_viridis_d(option = "plasma", begin = 0.1, end = 0.9) +
guides(fill = FALSE) +
labs(x = NULL, y = "Landscape fitness, linked",
caption = "Point = mean value") +
facet_wrap(~ constraint_clean, scales = "free_x") +
theme_bw()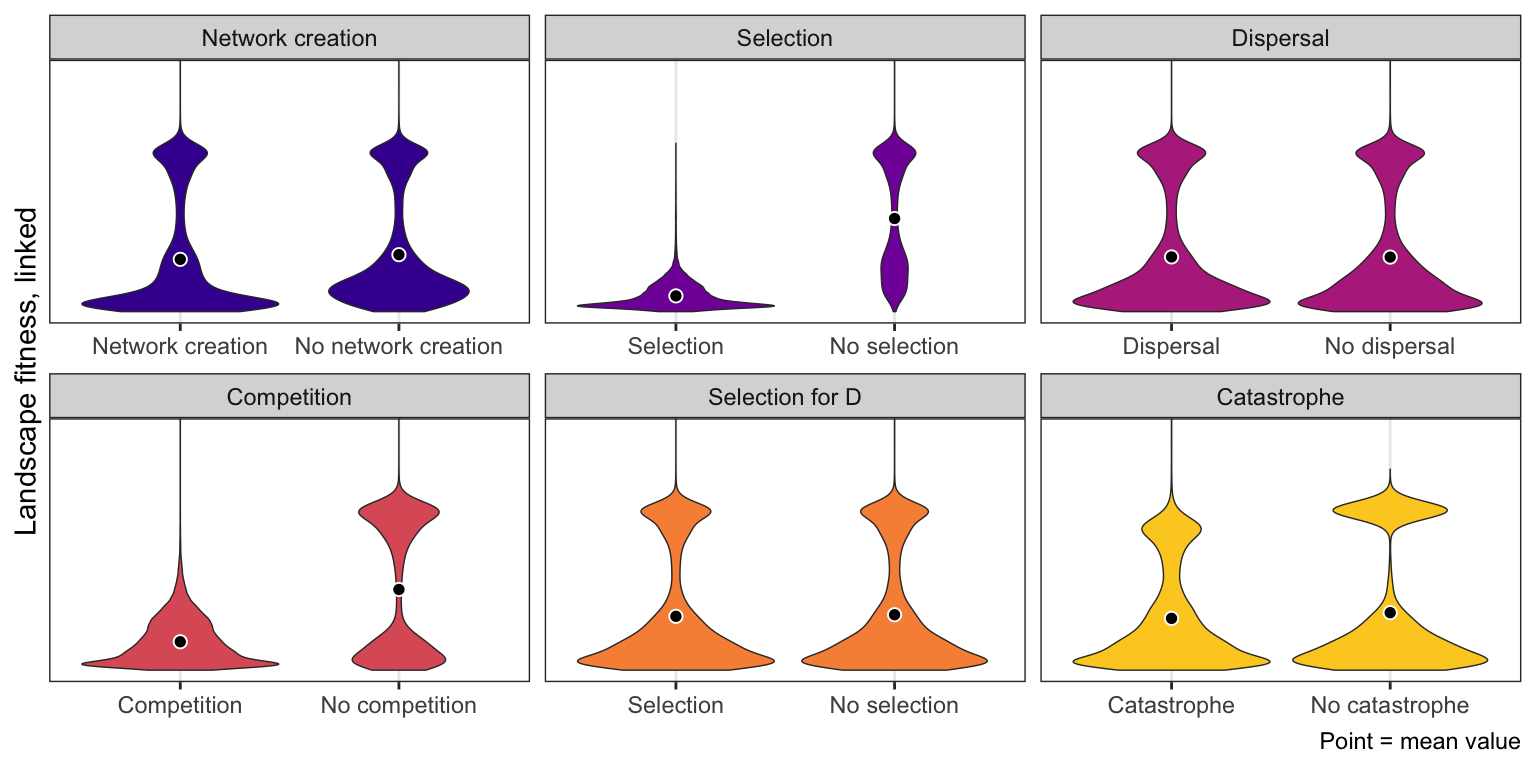
Nested interactions
Landscape fitness, linked
High-low
ggplot(BHL, aes(x = compete, y = landscape_fitness_linked)) +
geom_violin(aes(fill = repp), size = 0.1, width = 1,
position = position_dodge(width = 1)) +
stat_summary(aes(group = repp), geom = "point", fun = "mean",
position = position_dodge(width = 1),
size = 2, pch = 21, fill = "black", color = "white") +
scale_y_reverse(breaks = c(0, 0.2, 0.4)) +
coord_cartesian(ylim = c(0, 0.5)) +
labs(x = NULL, y = "Landscape fitness, linked",
fill = NULL, caption = "Point = mean value") +
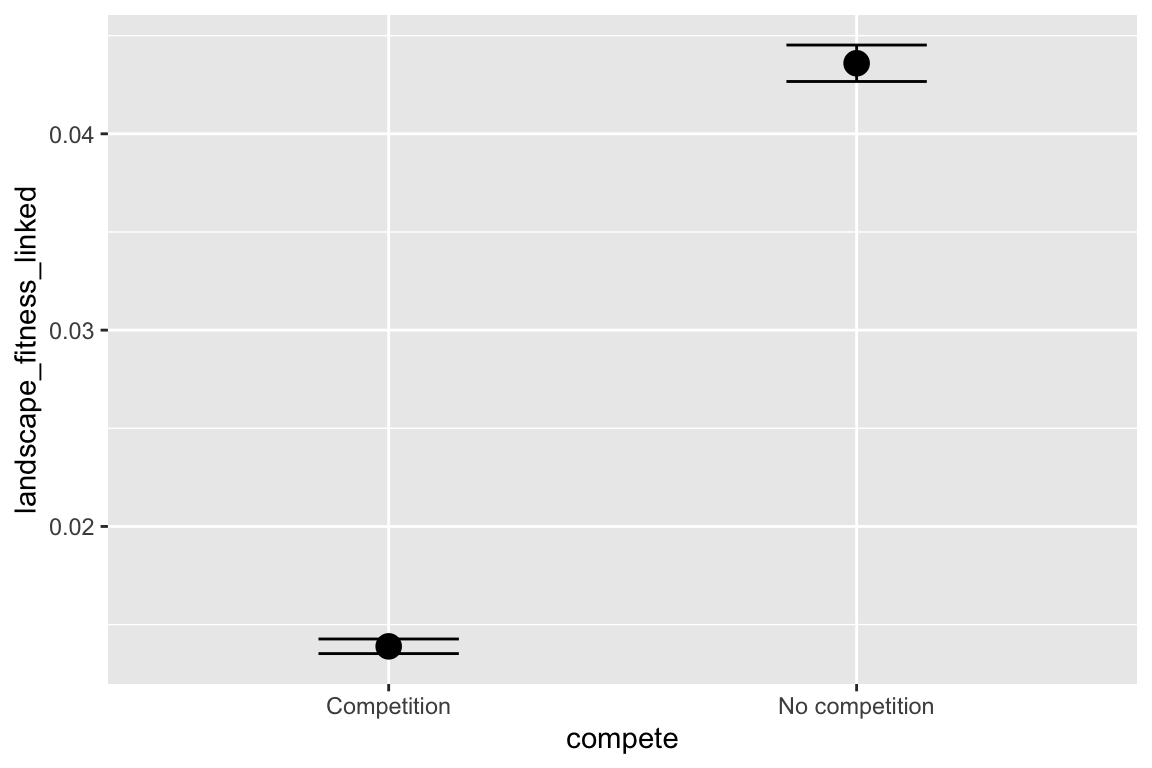
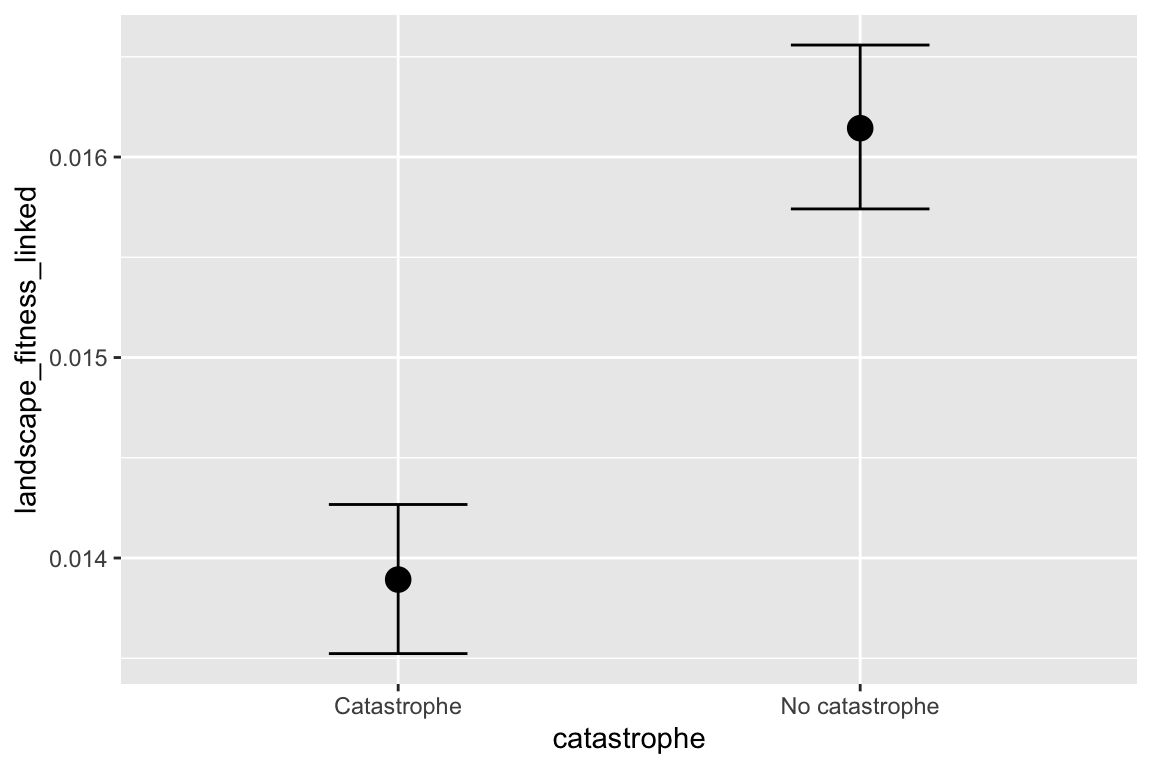
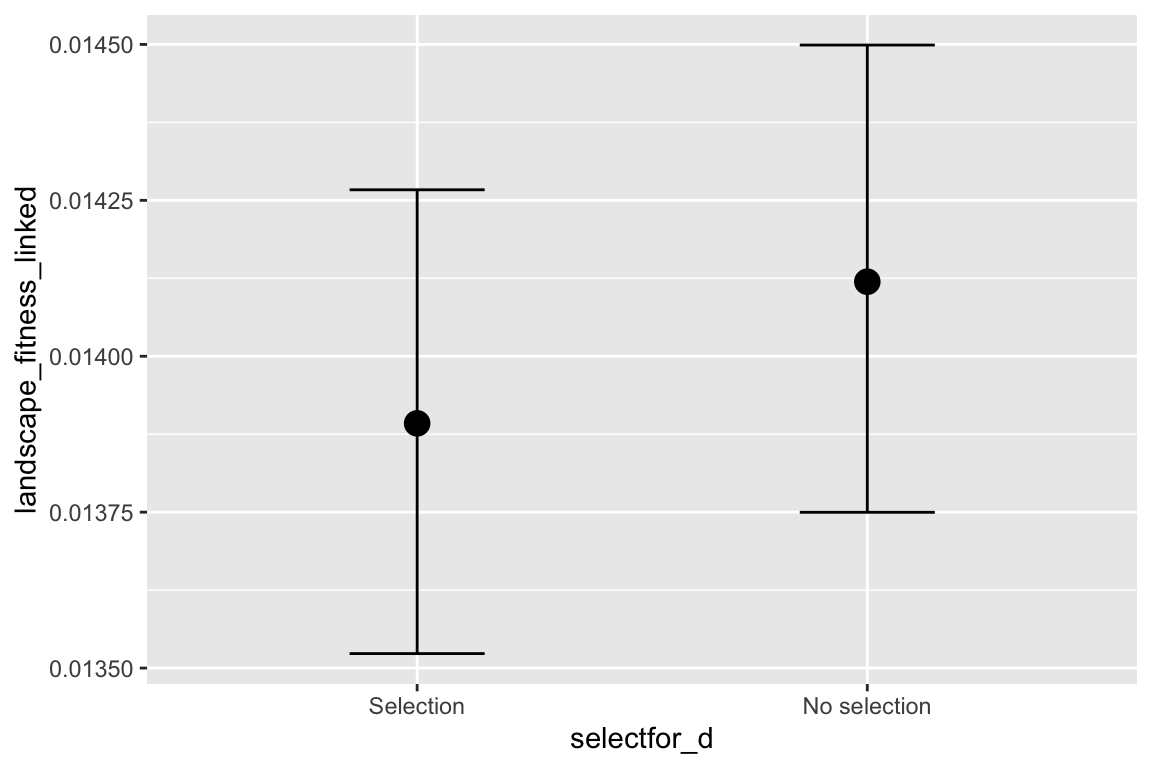
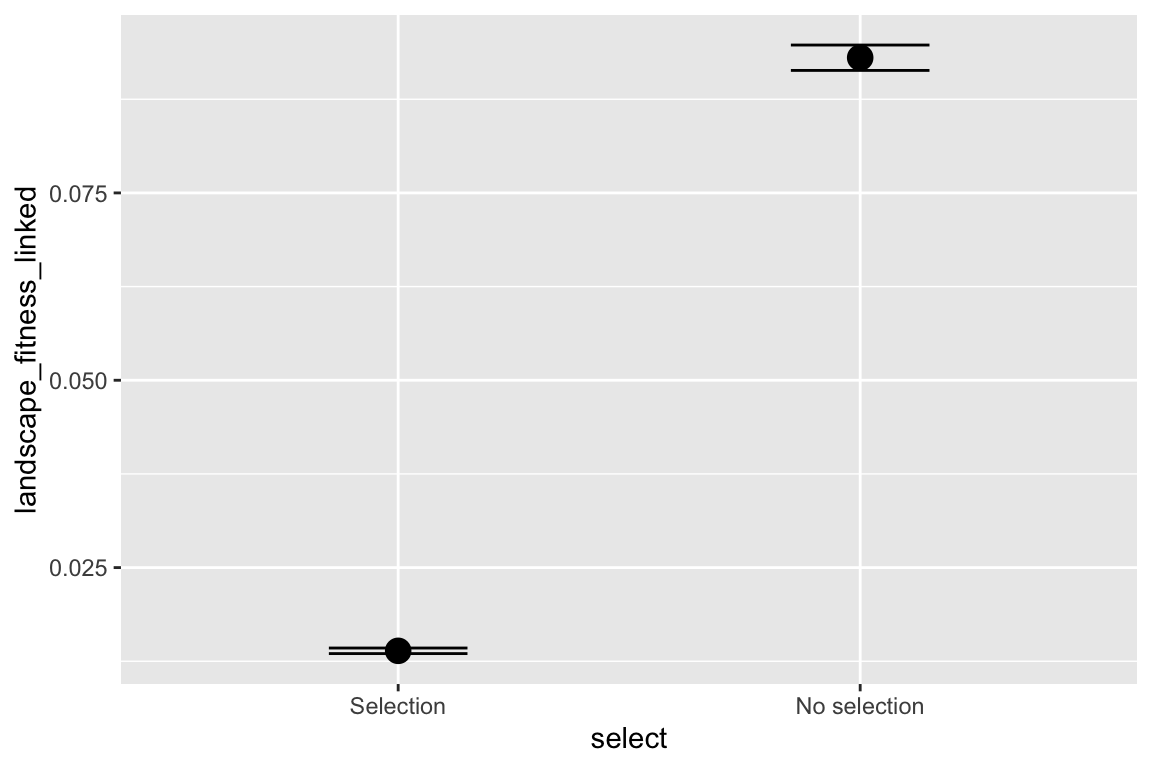
facet_grid(selectfor_d + create_network ~ disperse + catastrophe) +
theme_bw() +
theme(legend.position = "top",
legend.key.size = unit(0.65, "lines"))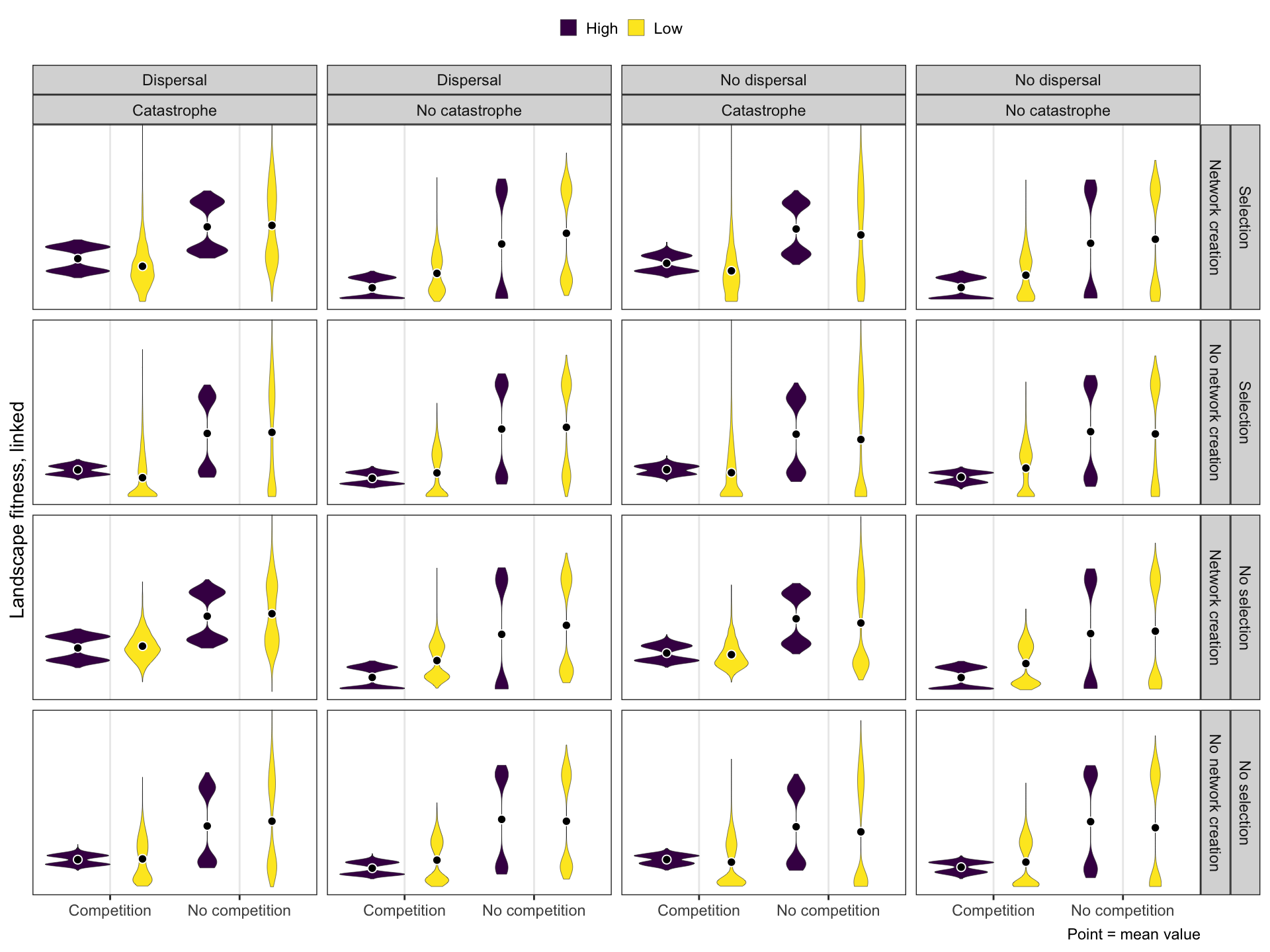
Latin squares
ggplot(BLS, aes(x = compete, y = landscape_fitness_linked)) +
geom_violin(fill = "#FE7F2D", size = 0.25) +
stat_summary(geom = "point", fun = "mean",
size = 2, pch = 21, fill = "black", color = "white") +
scale_y_reverse(breaks = c(0, 0.2, 0.4)) +
coord_cartesian(ylim = c(0, 0.5)) +
labs(x = NULL, y = "Landscape fitness, linked",
caption = "Point = mean value") +
facet_grid(selectfor_d + create_network ~ disperse + catastrophe) +
theme_bw()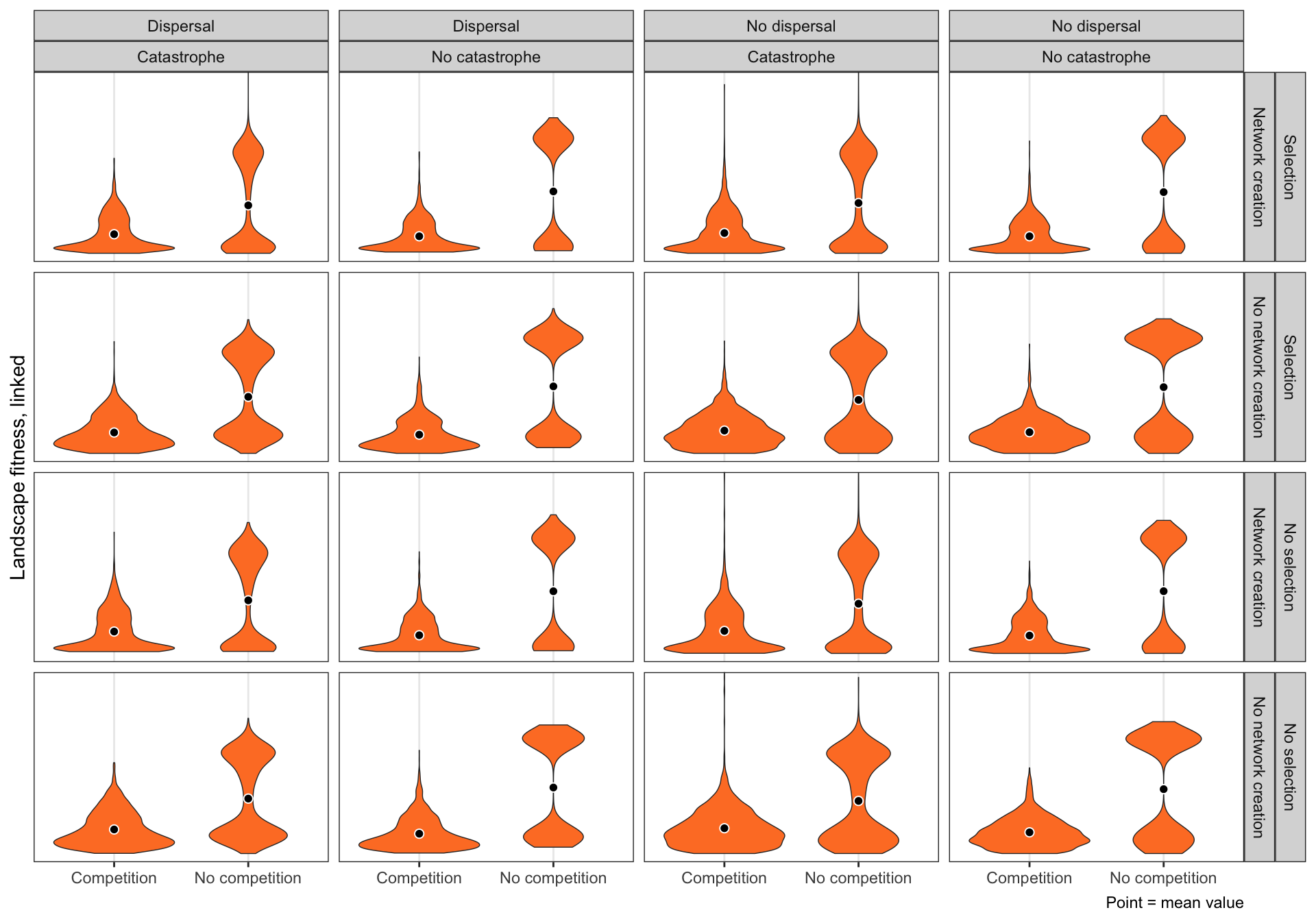
Modeling + marginal effects
Landscape fitness, linear model
model1 <- lm(landscape_fitness_linked ~ create_network + disperse + compete +
catastrophe + selectfor_d + select + numlink + cdis1 +
var_fitness + max_intial_proportion_links + maxsp,
data = BLS)
summary(model1)##
## Call:
## lm(formula = landscape_fitness_linked ~ create_network + disperse +
## compete + catastrophe + selectfor_d + select + numlink +
## cdis1 + var_fitness + max_intial_proportion_links + maxsp,
## data = BLS)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.28286 -0.05665 -0.00188 0.05300 0.43085
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -3.910e-02 1.210e-03 -32.307 < 2e-16 ***
## create_networkNo network creation 8.311e-03 4.950e-04 16.789 < 2e-16 ***
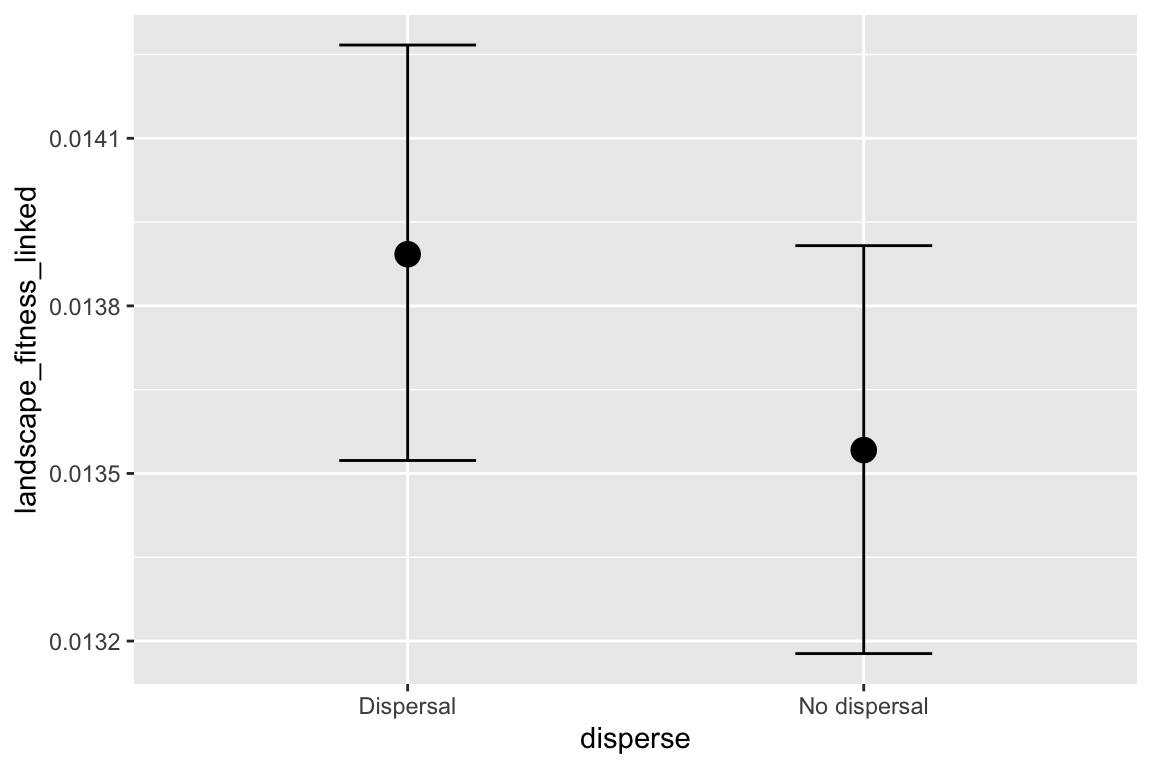
## disperseNo dispersal 3.427e-05 4.950e-04 0.069 0.9448
## competeNo competition 1.081e-01 4.950e-04 218.293 < 2e-16 ***
## catastropheNo catastrophe 1.335e-02 4.950e-04 26.965 < 2e-16 ***
## selectfor_dNo selection 2.361e-03 4.950e-04 4.769 1.86e-06 ***
## selectNo selection 1.608e-01 4.950e-04 324.736 < 2e-16 ***
## numlink 6.856e-04 1.396e-04 4.913 9.00e-07 ***
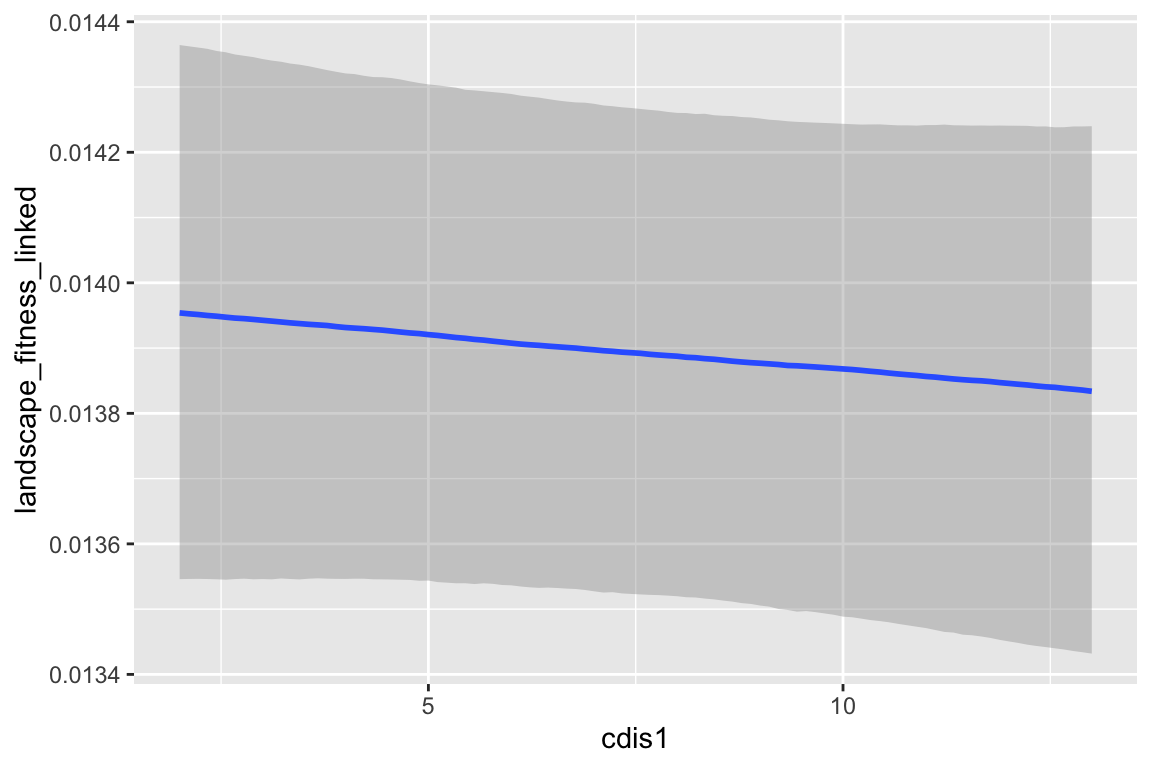
## cdis1 -1.237e-04 7.155e-05 -1.729 0.0838 .
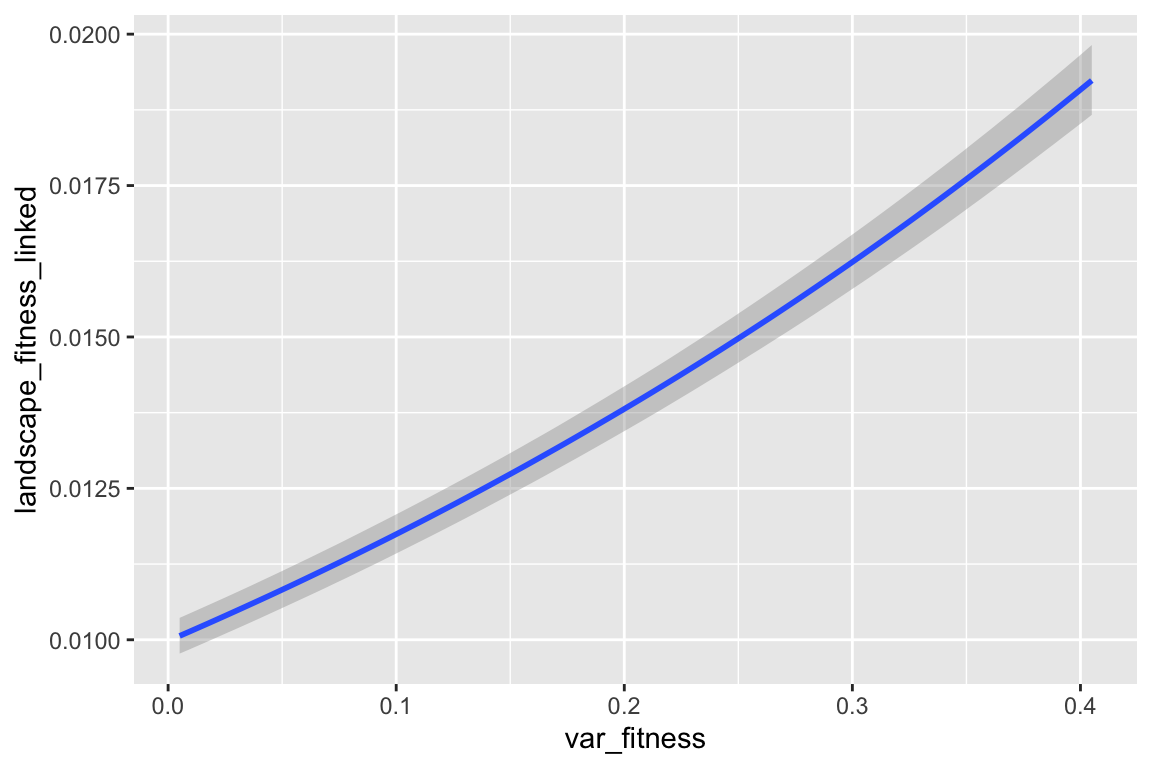
## var_fitness 1.328e-01 2.150e-03 61.771 < 2e-16 ***
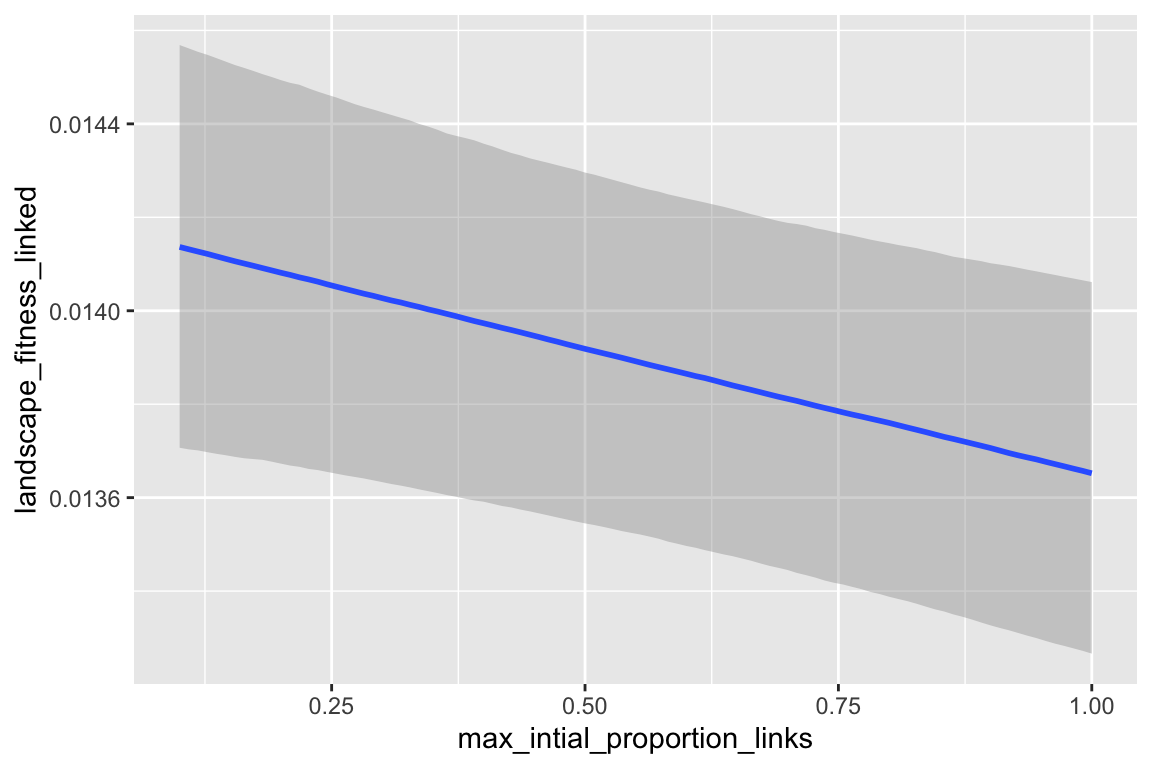
## max_intial_proportion_links -2.173e-04 9.540e-04 -0.228 0.8198
## maxsp -9.868e-04 5.001e-05 -19.734 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.06261 on 63988 degrees of freedom
## Multiple R-squared: 0.7151, Adjusted R-squared: 0.7151
## F-statistic: 1.46e+04 on 11 and 63988 DF, p-value: < 2.2e-16| (1) | |
| (Intercept) | -0.039 *** |
| (0.001) | |
| create_networkNo network creation | 0.008 *** |
| (0.000) | |
| disperseNo dispersal | 0.000 |
| (0.000) | |
| competeNo competition | 0.108 *** |
| (0.000) | |
| catastropheNo catastrophe | 0.013 *** |
| (0.000) | |
| selectfor_dNo selection | 0.002 *** |
| (0.000) | |
| selectNo selection | 0.161 *** |
| (0.000) | |
| numlink | 0.001 *** |
| (0.000) | |
| cdis1 | -0.000 |
| (0.000) | |
| var_fitness | 0.133 *** |
| (0.002) | |
| max_intial_proportion_links | -0.000 |
| (0.001) | |
| maxsp | -0.001 *** |
| (0.000) | |
| N | 64000 |
| R2 | 0.715 |
| logLik | 86527.063 |
| AIC | -173028.127 |
| *** p < 0.001; ** p < 0.01; * p < 0.05. | |
typical_values <- BLS %>%
summarize_all(typical) %>%
mutate(index = 1)
predictions1 <- crossing(numlink = 0:25, create_network = levels(BLS$create_network)) %>%
mutate(index = 1) %>%
left_join(select(typical_values, -numlink, -create_network), by = "index") %>%
select(-index) %>%
augment(model1, newdata = ., se_fit = TRUE) %>%
mutate(pred = .fitted,
pred.lower = pred + (qnorm(0.025) * .se.fit),
pred.upper = pred + (qnorm(0.975) * .se.fit))
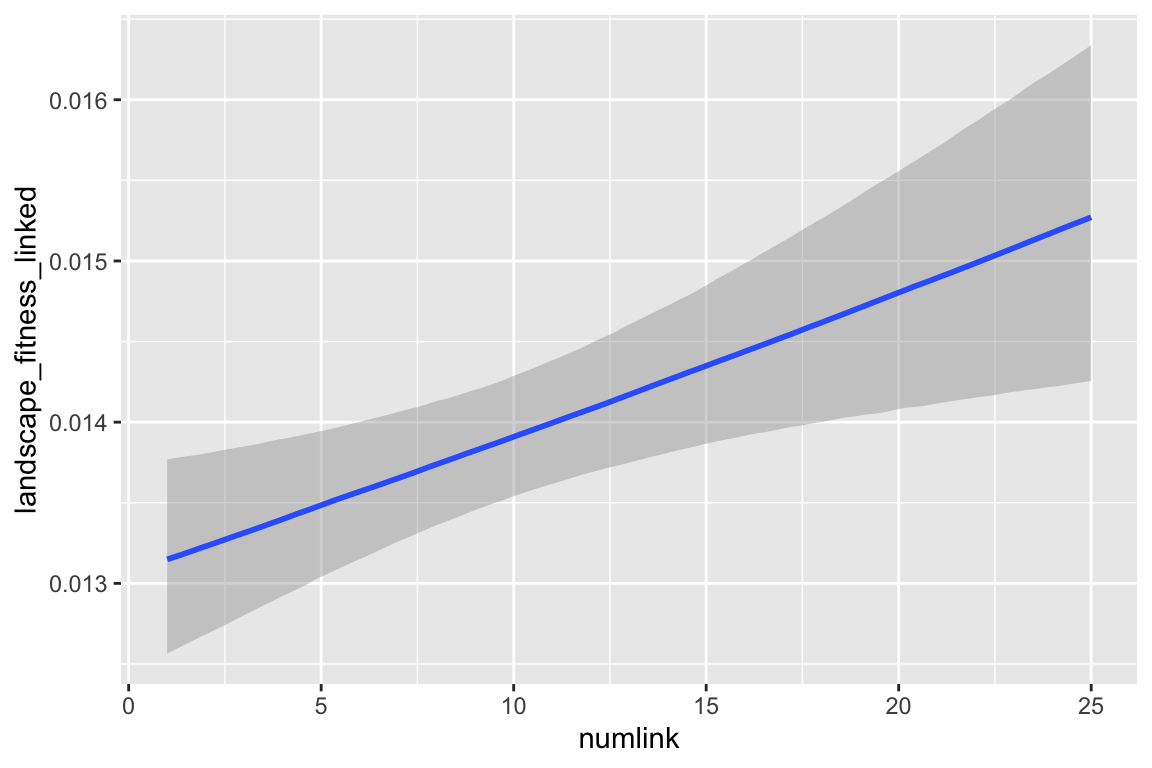
ggplot(predictions1, aes(x = numlink, y = pred, color = create_network)) +
geom_ribbon(aes(ymin = pred.lower, ymax = pred.upper, fill = create_network),
alpha = 0.3, color = NA) +
geom_line(size = 1.5) +
scale_y_reverse(labels = percent) +
scale_color_viridis_d(option = "plasma", end = 0.8) +
scale_fill_viridis_d(option = "plasma", end = 0.8) +
labs(y = "Predicted landscape fitness", x = "Number of links", color = NULL) +
guides(fill = FALSE) +
theme_bw() +
theme(legend.position = "bottom")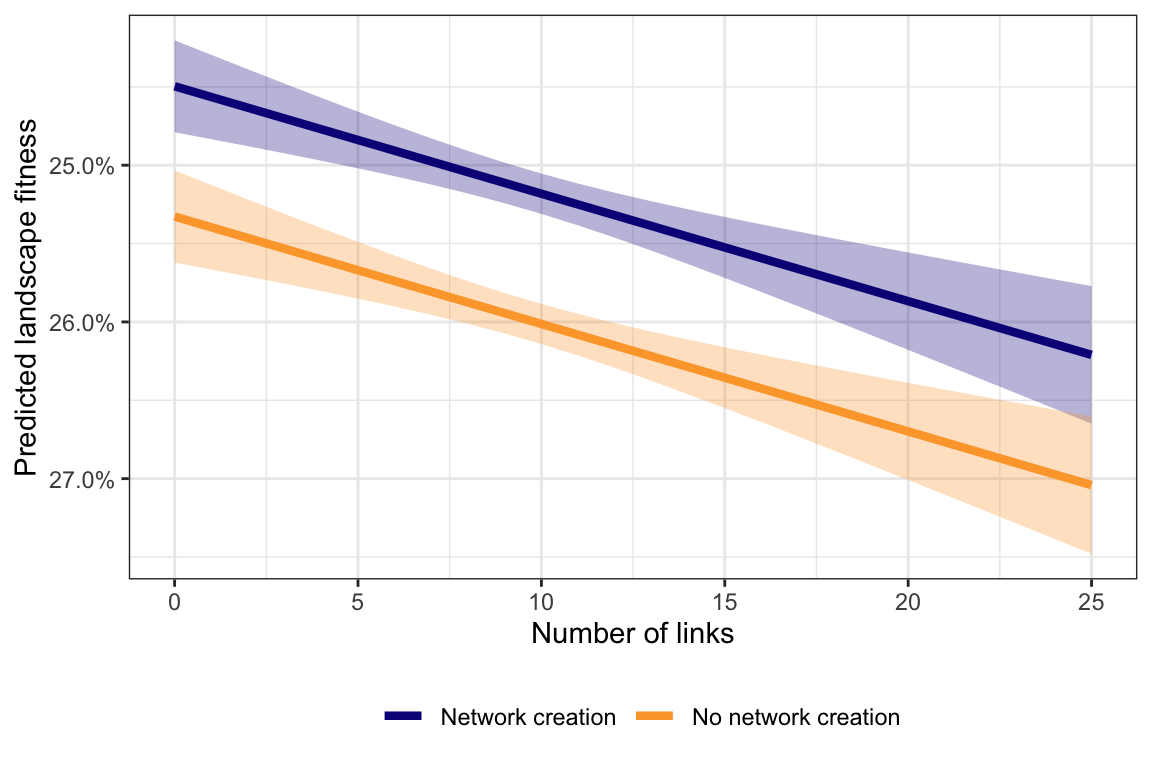
Landscape fitness, zero-inflated beta model
Histogram of landscape fitness to show zero-inflatedness + constraint within 0-1:
ggplot(BLS, aes(x = landscape_fitness_linked)) +
geom_histogram(bins = 100, color = "white") +
scale_x_continuous(labels = percent) +
theme_bw()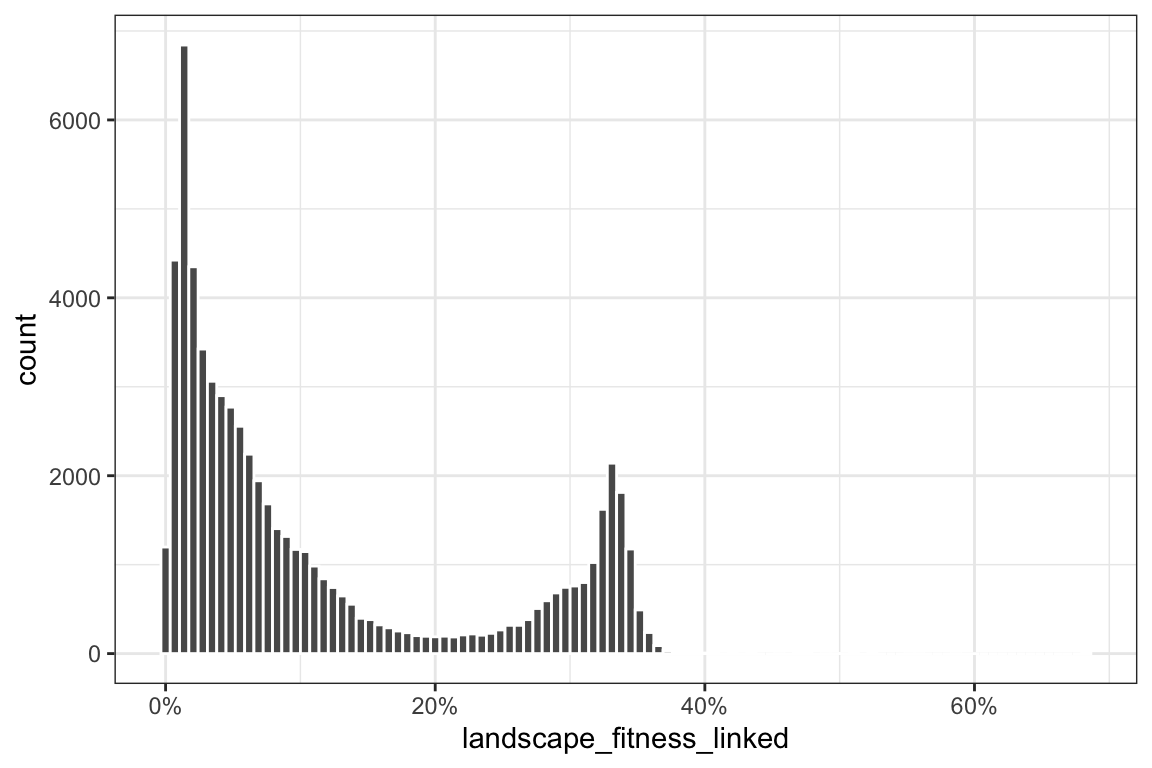
Build model
## Family: zero_inflated_beta
## Links: mu = logit; phi = identity; zi = identity
## Formula: landscape_fitness_linked ~ create_network + disperse + compete + catastrophe + selectfor_d + select + numlink + cdis1 + var_fitness + max_intial_proportion_links + maxsp
## Data: BLS_small (Number of observations: 16000)
## Samples: 4 chains, each with iter = 4000; warmup = 2000; thin = 1;
## total post-warmup samples = 8000
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Rhat
## Intercept -4.32 0.02 -4.36 -4.28 1.00
## create_networkNonetworkcreation 0.12 0.01 0.10 0.13 1.00
## disperseNodispersal -0.03 0.01 -0.04 -0.01 1.00
## competeNocompetition 1.17 0.01 1.16 1.19 1.00
## catastropheNocatastrophe 0.15 0.01 0.14 0.17 1.00
## selectfor_dNoselection 0.02 0.01 0.00 0.03 1.00
## selectNoselection 1.99 0.01 1.97 2.00 1.00
## numlink 0.01 0.00 0.00 0.01 1.00
## cdis1 -0.00 0.00 -0.00 0.00 1.00
## var_fitness 1.64 0.03 1.58 1.71 1.00
## max_intial_proportion_links -0.04 0.01 -0.07 -0.01 1.00
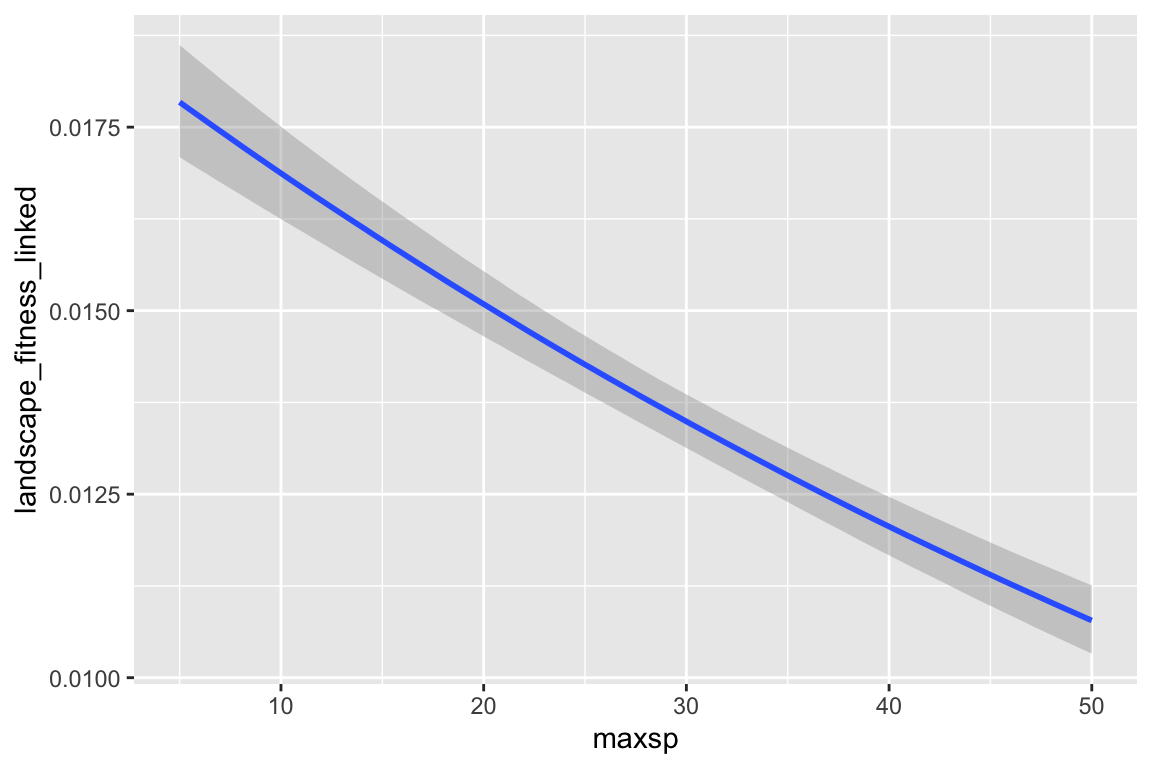
## maxsp -0.01 0.00 -0.01 -0.01 1.00
## Bulk_ESS Tail_ESS
## Intercept 7333 6271
## create_networkNonetworkcreation 10443 6134
## disperseNodispersal 10740 6122
## competeNocompetition 7064 6428
## catastropheNocatastrophe 9366 5392
## selectfor_dNoselection 10848 5493
## selectNoselection 5955 5751
## numlink 15260 4808
## cdis1 10038 4993
## var_fitness 8490 5657
## max_intial_proportion_links 10226 6012
## maxsp 13425 5594
##
## Family Specific Parameters:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## phi 46.89 0.55 45.81 47.99 1.00 6684 5976
## zi 0.00 0.00 0.00 0.00 1.00 9224 5416
##
## Samples were drawn using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).## Warning: Method 'marginal_effects' is deprecated. Please use
## 'conditional_effects' instead.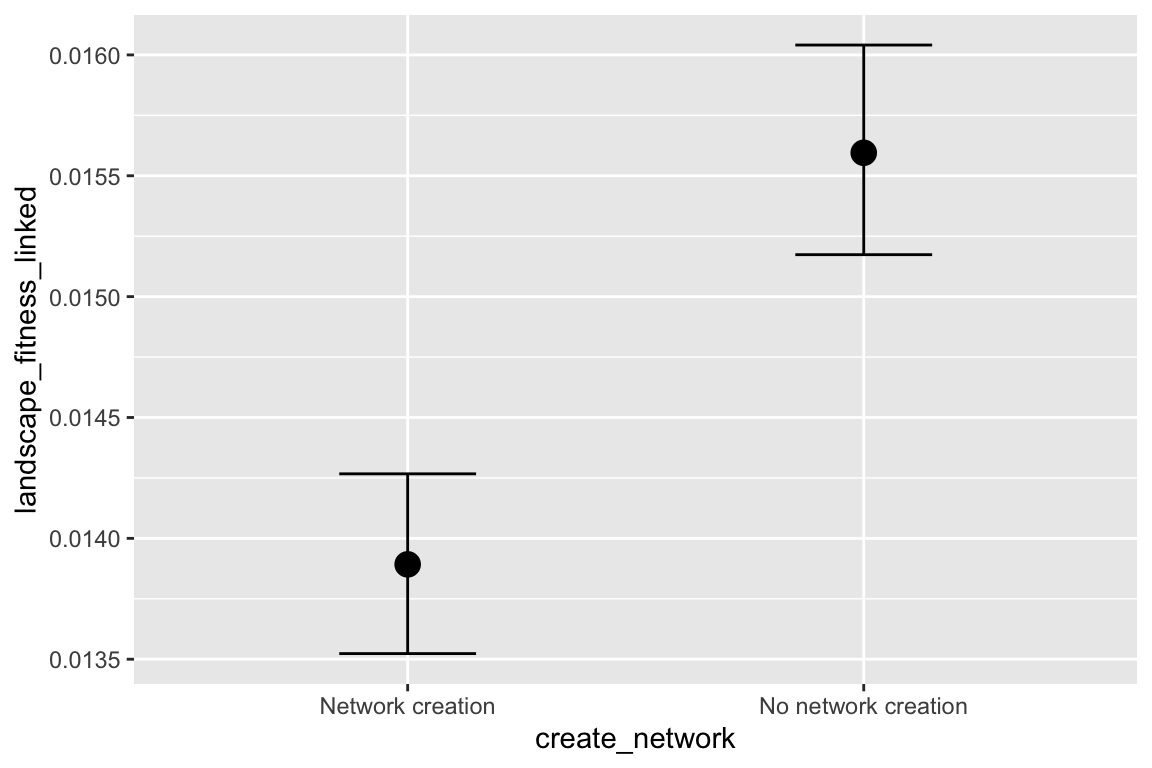
LS0tCnRpdGxlOiAiUHJlbGltaW5hcnkgYW5hbHlzaXMiCmF1dGhvcjogIlN0ZXZlbiBMLiBQZWNrIGFuZCBBbmRyZXcgSGVpc3MiCmRhdGU6ICJMYXN0IHJ1bjogYHIgZm9ybWF0KFN5cy50aW1lKCksICclQiAlZSwgJVknKWAiCm91dHB1dDogCiAgaHRtbF9kb2N1bWVudDoKICAgIGNvZGVfZm9sZGluZzogaGlkZQplZGl0b3Jfb3B0aW9uczogCiAgY2h1bmtfb3V0cHV0X3R5cGU6IGNvbnNvbGUKLS0tCgpgYGB7ciBsb2FkLWxpYnJhcmllcy1kYXRhLCB3YXJuaW5nPUZBTFNFLCBtZXNzYWdlPUZBTFNFfQpsaWJyYXJ5KHRpZHl2ZXJzZSkKbGlicmFyeShicm1zKQpsaWJyYXJ5KG1vZGVscikKbGlicmFyeShicm9vbSkKbGlicmFyeShodXh0YWJsZSkKbGlicmFyeShzY2FsZXMpCmxpYnJhcnkoaGVyZSkKCkNIQUlOUyA8LSA0CklURVIgPC0gNDAwMApXQVJNVVAgPC0gMjAwMApCQVlFU19TRUVEIDwtIDEyMzQKb3B0aW9ucyhtYy5jb3JlcyA9IHBhcmFsbGVsOjpkZXRlY3RDb3JlcygpKQoKY29uc3RyYWludHMgPC0gcmVhZF9yZHMoaGVyZSgiZGF0YSIsICJkZXJpdmVkX2RhdGEiLCAiY29uc3RyYWludHMucmRzIikpIAoKY29uc3RyYWludF9sZXZlbHMgPC0gY29uc3RyYWludHMgJT4lIAogIG11dGF0ZShjb25zdHJhaW50X2NsZWFuID0gZmN0X2lub3JkZXIoY29uc3RyYWludF9jbGVhbikpICU+JSAKICBtdXRhdGUobGV2ZWxzID0gbWFwKGxldmVsc19jbGVhbiwgbmFtZXMpKSAlPiUgCiAgdW5uZXN0KGxldmVscykgJT4lIAogIG11dGF0ZShsZXZlbHMgPSBmY3RfaW5vcmRlcihsZXZlbHMpKQoKIyBUT0RPOiBTd2l0Y2ggYmFjayB0byBmdWxsIEJMUy5yZHMgYW5kIEJITC5yZHMKQkhMIDwtIHJlYWRfcmRzKGhlcmUoImRhdGEiLCAiZGVyaXZlZF9kYXRhIiwgIkJITF9zbWFsbC5yZHMiKSkKYmhsX2xvbmcgPC0gQkhMICU+JSAKICBnYXRoZXIoY29uc3RyYWludCwgdmFsdWUsIG9uZV9vZihjb25zdHJhaW50cyRjb25zdHJhaW50KSkgJT4lIAogIGxlZnRfam9pbihjb25zdHJhaW50cywgYnkgPSAiY29uc3RyYWludCIpICU+JSAKICBtdXRhdGUodmFsdWUgPSBmY3RfcmVsZXZlbCh2YWx1ZSwgbGV2ZWxzKGNvbnN0cmFpbnRfbGV2ZWxzJGxldmVscykpLAogICAgICAgICBjb25zdHJhaW50X2NsZWFuID0gZmN0X3JlbGV2ZWwoY29uc3RyYWludF9jbGVhbiwgCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICBsZXZlbHMoY29uc3RyYWludF9sZXZlbHMkY29uc3RyYWludF9jbGVhbikpKQoKQkxTIDwtIHJlYWRfcmRzKGhlcmUoImRhdGEiLCAiZGVyaXZlZF9kYXRhIiwgIkJMU19zbWFsbC5yZHMiKSkKYmxzX2xvbmcgPC0gQkxTICU+JSAKICBnYXRoZXIoY29uc3RyYWludCwgdmFsdWUsIG9uZV9vZihjb25zdHJhaW50cyRjb25zdHJhaW50KSkgJT4lIAogIGxlZnRfam9pbihjb25zdHJhaW50cywgYnkgPSAiY29uc3RyYWludCIpICU+JSAKICBtdXRhdGUodmFsdWUgPSBmY3RfcmVsZXZlbCh2YWx1ZSwgbGV2ZWxzKGNvbnN0cmFpbnRfbGV2ZWxzJGxldmVscykpLAogICAgICAgICBjb25zdHJhaW50X2NsZWFuID0gZmN0X3JlbGV2ZWwoY29uc3RyYWludF9jbGVhbiwgCiAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICAgICBsZXZlbHMoY29uc3RyYWludF9sZXZlbHMkY29uc3RyYWludF9jbGVhbikpKQpgYGAKCgojIFNpbmdsZSBjb25kaXRpb25zLCBubyBuZXN0ZWQgaW50ZXJhY3Rpb25zCgojIyBMYW5kc2NhcGUgZml0bmVzcwoKIyMjIEhpZ2gtbG93CgpgYGB7ciBsYW5kc2NhcGUtZml0bmVzcy1saW5rZWQtYmhsLXNpbmdsZSwgZmlnLndpZHRoPTgsIGZpZy5oZWlnaHQ9NC41fQpnZ3Bsb3QoYmhsX2xvbmcsIGFlcyh4ID0gdmFsdWUsIHkgPSBhdmdfZXZlbm5lc3NfdCwgZmlsbCA9IHJlcHApKSArCiAgZ2VvbV92aW9saW4oc2l6ZSA9IDAuMjUsIHBvc2l0aW9uID0gcG9zaXRpb25fZG9kZ2Uod2lkdGggPSAwLjc1KSkgKwogIHN0YXRfc3VtbWFyeShnZW9tID0gInBvaW50IiwgZnVuID0gIm1lYW4iLCAKICAgICAgICAgICAgICAgYWVzKGdyb3VwID0gcmVwcCksIHBvc2l0aW9uID0gcG9zaXRpb25fZG9kZ2Uod2lkdGggPSAwLjc1KSwKICAgICAgICAgICAgICAgc2l6ZSA9IDIsIHBjaCA9IDIxLCBmaWxsID0gImJsYWNrIiwgY29sb3IgPSAid2hpdGUiKSArCiAgIyBzY2FsZV95X3JldmVyc2UoYnJlYWtzID0gc2VxKDAsIDAuNCwgMC4yKSkgKwogICMgY29vcmRfY2FydGVzaWFuKHlsaW0gPSBjKDAsIDAuNSkpICsKICBzY2FsZV9maWxsX3ZpcmlkaXNfZChvcHRpb24gPSAicGxhc21hIiwgYmVnaW4gPSAwLjEsIGVuZCA9IDAuOSkgKwogIGxhYnMoeCA9IE5VTEwsIHkgPSAiTGFuZHNjYXBlIGZpdG5lc3MsIGxpbmtlZCIsIGZpbGwgPSBOVUxMLCAKICAgICAgIGNhcHRpb24gPSAiUG9pbnQgPSBtZWFuIHZhbHVlIikgKwogIGZhY2V0X3dyYXAofiBjb25zdHJhaW50X2NsZWFuLCBzY2FsZXMgPSAiZnJlZV94IikgKwogIHRoZW1lX2J3KCkgKwogIHRoZW1lKGxlZ2VuZC5wb3NpdGlvbiA9ICJ0b3AiLAogICAgICAgIGxlZ2VuZC5rZXkuc2l6ZSA9IHVuaXQoMC42NSwgImxpbmVzIikpCmBgYAoKIyMjIExhdGluIHNxdWFyZXMKCmBgYHtyIGxhbmRzY2FwZS1maXRuZXNzLWxpbmtlZC1ibHMtc2luZ2xlLCBmaWcud2lkdGg9OCwgZmlnLmhlaWdodD00fQpnZ3Bsb3QoYmxzX2xvbmcsIAogICAgICAgYWVzKHggPSB2YWx1ZSwgeSA9IGxhbmRzY2FwZV9maXRuZXNzX2xpbmtlZCwgZmlsbCA9IGNvbnN0cmFpbnRfY2xlYW4pKSArCiAgZ2VvbV92aW9saW4oc2l6ZSA9IDAuMjUpICsKICBzdGF0X3N1bW1hcnkoZ2VvbSA9ICJwb2ludCIsIGZ1biA9ICJtZWFuIiwgCiAgICAgICAgICAgICAgIHNpemUgPSAyLCBwY2ggPSAyMSwgZmlsbCA9ICJibGFjayIsIGNvbG9yID0gIndoaXRlIikgKwogIHNjYWxlX3lfcmV2ZXJzZShicmVha3MgPSBzZXEoMCwgMC40LCAwLjIpKSArCiAgY29vcmRfY2FydGVzaWFuKHlsaW0gPSBjKDAsIDAuNSkpICsKICBzY2FsZV9maWxsX3ZpcmlkaXNfZChvcHRpb24gPSAicGxhc21hIiwgYmVnaW4gPSAwLjEsIGVuZCA9IDAuOSkgKwogIGd1aWRlcyhmaWxsID0gRkFMU0UpICsKICBsYWJzKHggPSBOVUxMLCB5ID0gIkxhbmRzY2FwZSBmaXRuZXNzLCBsaW5rZWQiLAogICAgICAgY2FwdGlvbiA9ICJQb2ludCA9IG1lYW4gdmFsdWUiKSArCiAgZmFjZXRfd3JhcCh+IGNvbnN0cmFpbnRfY2xlYW4sIHNjYWxlcyA9ICJmcmVlX3giKSArCiAgdGhlbWVfYncoKQpgYGAKCgojIE5lc3RlZCBpbnRlcmFjdGlvbnMKCiMjIExhbmRzY2FwZSBmaXRuZXNzLCBsaW5rZWQKCiMjIyBIaWdoLWxvdwoKYGBge3IgbGFuZHNjYXBlLWZpdG5lc3MtbGlua2VkLWJobCwgZmlnLndpZHRoPTEwLCBmaWcuaGVpZ2h0PTcuNX0KZ2dwbG90KEJITCwgYWVzKHggPSBjb21wZXRlLCB5ID0gbGFuZHNjYXBlX2ZpdG5lc3NfbGlua2VkKSkgKwogIGdlb21fdmlvbGluKGFlcyhmaWxsID0gcmVwcCksIHNpemUgPSAwLjEsIHdpZHRoID0gMSwKICAgICAgICAgICAgICBwb3NpdGlvbiA9IHBvc2l0aW9uX2RvZGdlKHdpZHRoID0gMSkpICsKICBzdGF0X3N1bW1hcnkoYWVzKGdyb3VwID0gcmVwcCksIGdlb20gPSAicG9pbnQiLCBmdW4gPSAibWVhbiIsIAogICAgICAgICAgICAgICBwb3NpdGlvbiA9IHBvc2l0aW9uX2RvZGdlKHdpZHRoID0gMSksCiAgICAgICAgICAgICAgIHNpemUgPSAyLCBwY2ggPSAyMSwgZmlsbCA9ICJibGFjayIsIGNvbG9yID0gIndoaXRlIikgKwogIHNjYWxlX3lfcmV2ZXJzZShicmVha3MgPSBjKDAsIDAuMiwgMC40KSkgKwogIGNvb3JkX2NhcnRlc2lhbih5bGltID0gYygwLCAwLjUpKSArCiAgbGFicyh4ID0gTlVMTCwgeSA9ICJMYW5kc2NhcGUgZml0bmVzcywgbGlua2VkIiwgCiAgICAgICBmaWxsID0gTlVMTCwgY2FwdGlvbiA9ICJQb2ludCA9IG1lYW4gdmFsdWUiKSArCiAgZmFjZXRfZ3JpZChzZWxlY3Rmb3JfZCArIGNyZWF0ZV9uZXR3b3JrIH4gZGlzcGVyc2UgKyBjYXRhc3Ryb3BoZSkgKwogIHRoZW1lX2J3KCkgKwogIHRoZW1lKGxlZ2VuZC5wb3NpdGlvbiA9ICJ0b3AiLAogICAgICAgIGxlZ2VuZC5rZXkuc2l6ZSA9IHVuaXQoMC42NSwgImxpbmVzIikpCmBgYAoKIyMjIExhdGluIHNxdWFyZXMKCmBgYHtyIGxhbmRzY2FwZS1maXRuZXNzLWxpbmtlZC1ibHMsIGZpZy53aWR0aD0xMCwgZmlnLmhlaWdodD03fQpnZ3Bsb3QoQkxTLCBhZXMoeCA9IGNvbXBldGUsIHkgPSBsYW5kc2NhcGVfZml0bmVzc19saW5rZWQpKSArCiAgZ2VvbV92aW9saW4oZmlsbCA9ICIjRkU3RjJEIiwgc2l6ZSA9IDAuMjUpICsKICBzdGF0X3N1bW1hcnkoZ2VvbSA9ICJwb2ludCIsIGZ1biA9ICJtZWFuIiwgCiAgICAgICAgICAgICAgIHNpemUgPSAyLCBwY2ggPSAyMSwgZmlsbCA9ICJibGFjayIsIGNvbG9yID0gIndoaXRlIikgKwogIHNjYWxlX3lfcmV2ZXJzZShicmVha3MgPSBjKDAsIDAuMiwgMC40KSkgKwogIGNvb3JkX2NhcnRlc2lhbih5bGltID0gYygwLCAwLjUpKSArCiAgbGFicyh4ID0gTlVMTCwgeSA9ICJMYW5kc2NhcGUgZml0bmVzcywgbGlua2VkIiwKICAgICAgIGNhcHRpb24gPSAiUG9pbnQgPSBtZWFuIHZhbHVlIikgKwogIGZhY2V0X2dyaWQoc2VsZWN0Zm9yX2QgKyBjcmVhdGVfbmV0d29yayB+IGRpc3BlcnNlICsgY2F0YXN0cm9waGUpICsKICB0aGVtZV9idygpCmBgYAoKCiMgTW9kZWxpbmcgKyBtYXJnaW5hbCBlZmZlY3RzCgojIyBMYW5kc2NhcGUgZml0bmVzcywgbGluZWFyIG1vZGVsCgpgYGB7ciBtb2RlbC1sYW5kc2NhcGUtZml0bmVzc30KbW9kZWwxIDwtIGxtKGxhbmRzY2FwZV9maXRuZXNzX2xpbmtlZCB+IGNyZWF0ZV9uZXR3b3JrICsgZGlzcGVyc2UgKyBjb21wZXRlICsgCiAgICAgICAgICAgICAgIGNhdGFzdHJvcGhlICsgc2VsZWN0Zm9yX2QgKyBzZWxlY3QgKyBudW1saW5rICsgY2RpczEgKyAKICAgICAgICAgICAgICAgdmFyX2ZpdG5lc3MgKyBtYXhfaW50aWFsX3Byb3BvcnRpb25fbGlua3MgKyBtYXhzcCwKICAgICAgICAgICAgIGRhdGEgPSBCTFMpCnN1bW1hcnkobW9kZWwxKQpodXhyZWcobW9kZWwxKQpgYGAKCmBgYHtyIG1hcmdpbmFsLWVmZmVjdHMtbGFuZHNjYXBlLWZpdG5lc3N9CnR5cGljYWxfdmFsdWVzIDwtIEJMUyAlPiUgCiAgc3VtbWFyaXplX2FsbCh0eXBpY2FsKSAlPiUgCiAgbXV0YXRlKGluZGV4ID0gMSkKCnByZWRpY3Rpb25zMSA8LSBjcm9zc2luZyhudW1saW5rID0gMDoyNSwgY3JlYXRlX25ldHdvcmsgPSBsZXZlbHMoQkxTJGNyZWF0ZV9uZXR3b3JrKSkgJT4lIAogIG11dGF0ZShpbmRleCA9IDEpICU+JQogIGxlZnRfam9pbihzZWxlY3QodHlwaWNhbF92YWx1ZXMsIC1udW1saW5rLCAtY3JlYXRlX25ldHdvcmspLCBieSA9ICJpbmRleCIpICU+JQogIHNlbGVjdCgtaW5kZXgpICU+JQogIGF1Z21lbnQobW9kZWwxLCBuZXdkYXRhID0gLiwgc2VfZml0ID0gVFJVRSkgJT4lIAogIG11dGF0ZShwcmVkID0gLmZpdHRlZCwKICAgICAgICAgcHJlZC5sb3dlciA9IHByZWQgKyAocW5vcm0oMC4wMjUpICogLnNlLmZpdCksCiAgICAgICAgIHByZWQudXBwZXIgPSBwcmVkICsgKHFub3JtKDAuOTc1KSAqIC5zZS5maXQpKQoKZ2dwbG90KHByZWRpY3Rpb25zMSwgYWVzKHggPSBudW1saW5rLCB5ID0gcHJlZCwgY29sb3IgPSBjcmVhdGVfbmV0d29yaykpICsKICBnZW9tX3JpYmJvbihhZXMoeW1pbiA9IHByZWQubG93ZXIsIHltYXggPSBwcmVkLnVwcGVyLCBmaWxsID0gY3JlYXRlX25ldHdvcmspLAogICAgICAgICAgICAgIGFscGhhID0gMC4zLCBjb2xvciA9IE5BKSArCiAgZ2VvbV9saW5lKHNpemUgPSAxLjUpICsgCiAgc2NhbGVfeV9yZXZlcnNlKGxhYmVscyA9IHBlcmNlbnQpICsKICBzY2FsZV9jb2xvcl92aXJpZGlzX2Qob3B0aW9uID0gInBsYXNtYSIsIGVuZCA9IDAuOCkgKwogIHNjYWxlX2ZpbGxfdmlyaWRpc19kKG9wdGlvbiA9ICJwbGFzbWEiLCBlbmQgPSAwLjgpICsKICBsYWJzKHkgPSAiUHJlZGljdGVkIGxhbmRzY2FwZSBmaXRuZXNzIiwgeCA9ICJOdW1iZXIgb2YgbGlua3MiLCBjb2xvciA9IE5VTEwpICsgCiAgZ3VpZGVzKGZpbGwgPSBGQUxTRSkgKwogIHRoZW1lX2J3KCkgKwogIHRoZW1lKGxlZ2VuZC5wb3NpdGlvbiA9ICJib3R0b20iKQpgYGAKCiMjIExhbmRzY2FwZSBmaXRuZXNzLCB6ZXJvLWluZmxhdGVkIGJldGEgbW9kZWwKCkhpc3RvZ3JhbSBvZiBsYW5kc2NhcGUgZml0bmVzcyB0byBzaG93IHplcm8taW5mbGF0ZWRuZXNzICsgY29uc3RyYWludCB3aXRoaW4gMC0xOgoKYGBge3IgZml0bmVzcy1oaXN0fQpnZ3Bsb3QoQkxTLCBhZXMoeCA9IGxhbmRzY2FwZV9maXRuZXNzX2xpbmtlZCkpICsKICBnZW9tX2hpc3RvZ3JhbShiaW5zID0gMTAwLCBjb2xvciA9ICJ3aGl0ZSIpICsKICBzY2FsZV94X2NvbnRpbnVvdXMobGFiZWxzID0gcGVyY2VudCkgKwogIHRoZW1lX2J3KCkKYGBgCgpCdWlsZCBtb2RlbAoKYGBge3J9Cm1vZGVsX3ppbmIgPC0gcmVhZFJEUyhoZXJlKCJkYXRhIiwgImRlcml2ZWRfZGF0YSIsICJtb2RlbF96aW5iX3Rlc3QucmRzIikpCgpzdW1tYXJ5KG1vZGVsX3ppbmIpCgpwbG90KG1hcmdpbmFsX2VmZmVjdHMobW9kZWxfemluYikpCmBgYAo=