Additional analysis
Suparna Chaudhry and Andrew Heiss
Last run: 2020-10-01
# Load libraries
library(tidyverse)
library(crackdownsphilanthropy)
library(magrittr)
library(rstan)
library(rstanarm)
library(broom)
library(broom.mixed)
library(glue)
library(grid)
library(gridExtra)
library(Gmisc) # For the CONSORT diagram
library(pander)
library(scales)
library(ipumsr)
library(huxtable)
library(here)
source(here("analysis", "options.R"))
# Print correct huxtable table depending on the type of output.
#
# Technically this isn't completely necessary, since huxtable can output a
# markdown table, which is ostensibly universal for all output types. However,
# markdown tables are inherently limited in how fancy they can be (e.g. they
# don't support column spans), so I instead let the regression table use
# huxtable's fancy formatting for html and PDF and markdown everywhere else.
if (isTRUE(getOption('knitr.in.progress'))) {
file_format <- rmarkdown::all_output_formats(knitr::current_input())
} else {
file_format <- ""
}
print_hux <- function(x) {
if ("html_document" %in% file_format) {
print_html(x)
} else if ("pdf_document" %in% file_format) {
print_latex(x)
} else if ("word_document" %in% file_format) {
print_md(x)
} else {
print(x)
}
}
# Load data
results <- readRDS(here("data", "derived_data", "results_clean.rds"))CONSORT flow
consort <- readRDS(here("data", "derived_data", "completion_summary.rds")) %>%
pivot_wider(names_from = reason, values_from = n) %>%
mutate(group = 1:n(),
assigned = Approved + `Failed first attention check`,
issue = str_replace_all(issue, "assistance", "assist.")) %>%
mutate(assigned_label = glue("Allocated to Group {group}\n{crackdown}\n{issue}\n{funding} funding\n\nN = {assigned}"),
completed_label = glue("Completed\nN = {Approved}\n\n{`Failed first attention check`} failed\nattention check"))
assessed_eligibility_n <- sum(consort$Approved, consort$`Failed first attention check`,
consort$`Took survey outisde of MTurk`)
ineligible_n <- sum(consort$`Took survey outisde of MTurk`)
randomized_n <- sum(consort$Approved, consort$`Failed first attention check`)
# https://aghaynes.wordpress.com/2018/05/09/flow-charts-in-r/
# set some parameters to use repeatedly
width <- 0.1
xs <- seq(0.1, 0.9, length.out = 8)
allocated_y <- 0.375
completed_y <- 0.125
box_gp_grey <- gpar(fill = ngo_cols("light grey"))
box_gp_blue_dk <- gpar(fill = ngo_cols("blue"), alpha = 0.75)
box_gp_blue_lt <- gpar(fill = ngo_cols("blue"), alpha = 0.35)
box_gp_green <- gpar(fill = ngo_cols("green"), alpha = 0.65)
box_gp_yellow <- gpar(fill = ngo_cols("yellow"))
box_gp_orange <- gpar(fill = ngo_cols("orange"), alpha = 0.65)
txt_gp <- gpar(fontfamily = "Roboto Condensed",
fontface = "plain", fontsize = 8)
# Create boxes
total <- boxGrob(glue("Assessed for eligibility\n N = {assessed_eligibility_n}"),
x = 0.5, y = 0.9, width = 2 * width,
box_gp = box_gp_blue_lt, txt_gp = txt_gp)
randomized <- boxGrob(glue("Randomized\n N = {randomized_n}"),
x = 0.5, y = 0.65, width = 2 * width,
box_gp = box_gp_blue_dk, txt_gp = txt_gp)
ineligible <- boxGrob(glue("Participants excluded for\ncompleting Qualtrics survey\noutside of MTurk\n N = {ineligible_n}"),
x = xs[7], y = 0.775, #width = 0.25,
box_gp = box_gp_yellow, txt_gp = txt_gp)
group1 <- boxGrob(filter(consort, group == 1)$assigned_label,
x = xs[1], y = allocated_y, width = width,
box_gp = box_gp_orange, txt_gp = txt_gp)
group2 <- boxGrob(filter(consort, group == 2)$assigned_label,
x = xs[2], y = allocated_y, width = width,
box_gp = box_gp_orange, txt_gp = txt_gp)
group3 <- boxGrob(filter(consort, group == 3)$assigned_label,
x = xs[3], y = allocated_y, width = width,
box_gp = box_gp_orange, txt_gp = txt_gp)
group4 <- boxGrob(filter(consort, group == 4)$assigned_label,
x = xs[4], y = allocated_y, width = width,
box_gp = box_gp_orange, txt_gp = txt_gp)
group5 <- boxGrob(filter(consort, group == 5)$assigned_label,
x = xs[5], y = allocated_y, width = width,
box_gp = box_gp_orange, txt_gp = txt_gp)
group6 <- boxGrob(filter(consort, group == 6)$assigned_label,
x = xs[6], y = allocated_y, width = width,
box_gp = box_gp_orange, txt_gp = txt_gp)
group7 <- boxGrob(filter(consort, group == 7)$assigned_label,
x = xs[7], y = allocated_y, width = width,
box_gp = box_gp_orange, txt_gp = txt_gp)
group8 <- boxGrob(filter(consort, group == 8)$assigned_label,
x = xs[8], y = allocated_y, width = width,
box_gp = box_gp_orange, txt_gp = txt_gp)
group1_completed <- boxGrob(filter(consort, group == 1)$completed_label,
x = xs[1], y = completed_y, width = width,
box_gp = box_gp_green, txt_gp = txt_gp)
group2_completed <- boxGrob(filter(consort, group == 2)$completed_label,
x = xs[2], y = completed_y, width = width,
box_gp = box_gp_green, txt_gp = txt_gp)
group3_completed <- boxGrob(filter(consort, group == 3)$completed_label,
x = xs[3], y = completed_y, width = width,
box_gp = box_gp_green, txt_gp = txt_gp)
group4_completed <- boxGrob(filter(consort, group == 4)$completed_label,
x = xs[4], y = completed_y, width = width,
box_gp = box_gp_green, txt_gp = txt_gp)
group5_completed <- boxGrob(filter(consort, group == 5)$completed_label,
x = xs[5], y = completed_y, width = width,
box_gp = box_gp_green, txt_gp = txt_gp)
group6_completed <- boxGrob(filter(consort, group == 6)$completed_label,
x = xs[6], y = completed_y, width = width,
box_gp = box_gp_green, txt_gp = txt_gp)
group7_completed <- boxGrob(filter(consort, group == 7)$completed_label,
x = xs[7], y = completed_y, width = width,
box_gp = box_gp_green, txt_gp = txt_gp)
group8_completed <- boxGrob(filter(consort, group == 8)$completed_label,
x = xs[8], y = completed_y, width = width,
box_gp = box_gp_green, txt_gp = txt_gp)
total_random_connect <- connectGrob(total, randomized, "v")
total_ineligible_connect <- connectGrob(total, ineligible, "-")
rand_connect1 <- connectGrob(randomized, group1, "N")
rand_connect2 <- connectGrob(randomized, group2, "N")
rand_connect3 <- connectGrob(randomized, group3, "N")
rand_connect4 <- connectGrob(randomized, group4, "N")
rand_connect5 <- connectGrob(randomized, group5, "N")
rand_connect6 <- connectGrob(randomized, group6, "N")
rand_connect7 <- connectGrob(randomized, group7, "N")
rand_connect8 <- connectGrob(randomized, group8, "N")
complete_connect1 <- connectGrob(group1, group1_completed, "N")
complete_connect2 <- connectGrob(group2, group2_completed, "N")
complete_connect3 <- connectGrob(group3, group3_completed, "N")
complete_connect4 <- connectGrob(group4, group4_completed, "N")
complete_connect5 <- connectGrob(group5, group5_completed, "N")
complete_connect6 <- connectGrob(group6, group6_completed, "N")
complete_connect7 <- connectGrob(group7, group7_completed, "N")
complete_connect8 <- connectGrob(group8, group8_completed, "N")
full_chart <- list(total, randomized, ineligible, total_random_connect, total_ineligible_connect,
group1, group2, group3, group4, group5, group6, group7, group8,
rand_connect1, rand_connect2, rand_connect3, rand_connect4,
rand_connect5, rand_connect6, rand_connect7, rand_connect8,
group1_completed, group2_completed, group3_completed, group4_completed,
group5_completed, group6_completed, group7_completed, group8_completed,
complete_connect1, complete_connect2, complete_connect3, complete_connect4,
complete_connect5, complete_connect6, complete_connect7, complete_connect8) # Ordinarily, you can use grid.grab() to save the output of a grid figure into
# an object and then use that in ggsave(). However, when knitting, this creates
# a duplicate plot, which is frustrating. So instead, we use walk() to reprint
# all the grobs within specific pdf and png devices
#
# See https://stackoverflow.com/a/17509770/120898 for a similar issue
# Save as PDF
cairo_pdf(filename = here("analysis", "output", "figures", "consort.pdf"),
width = 10, height = 6)
grid.newpage()
walk(full_chart, ~ print(.))
invisible(dev.off())
# Save as PNG
png(filename = here("analysis", "output", "figures", "consort.png"),
width = 10, height = 6, units = "in",
bg = "white", res = 300, type = "cairo")
grid.newpage()
walk(full_chart, ~ print(.))
invisible(dev.off())
# Show in knitted document
grid.newpage()
walk(full_chart, ~ print(.))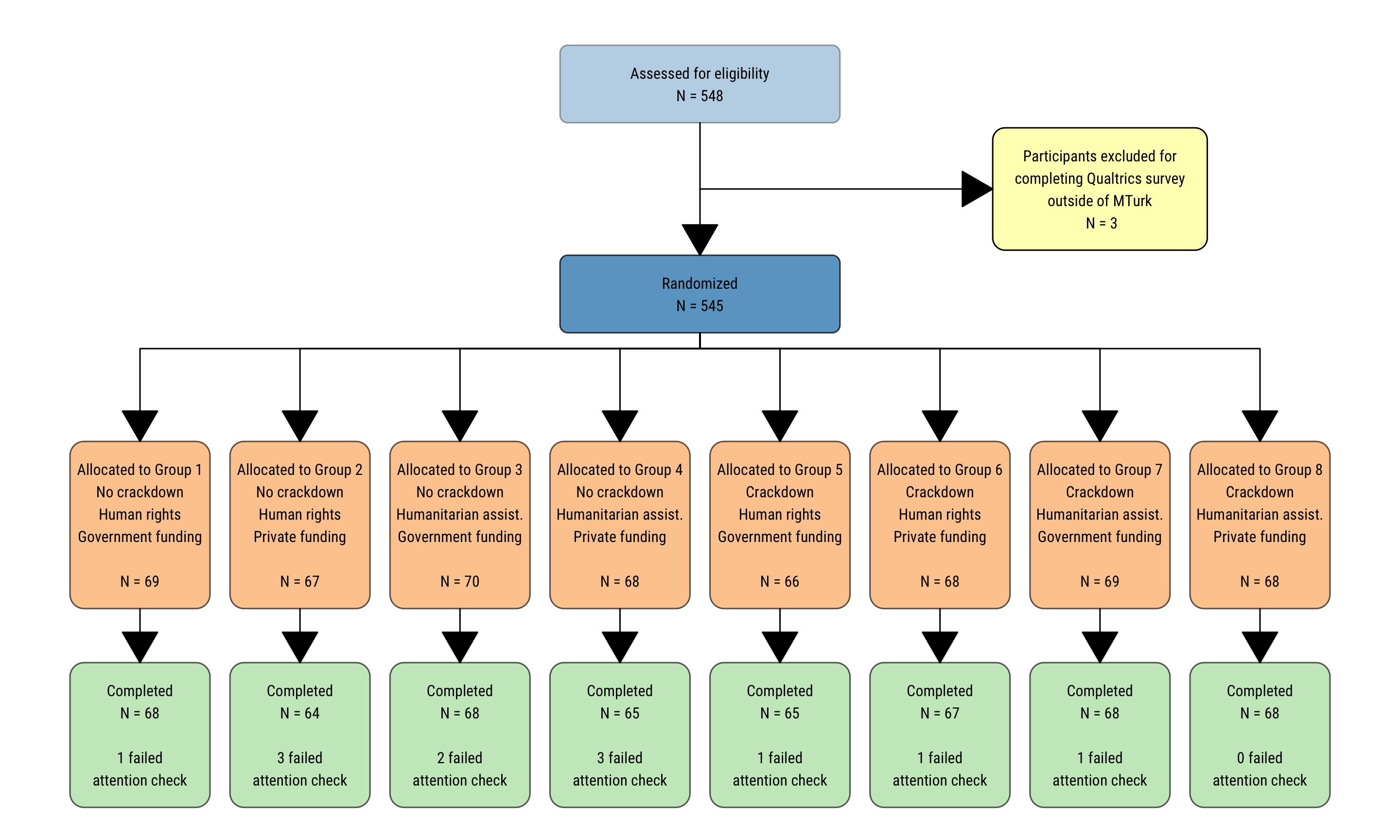
Characteristics of experiment samples
We compare our sample with demographic characteristics of the general population. Since there is no nationally representative sample for each of our demographic variables, we use two waves of the US Census’s Current Population Survey (CPS), with data from the Minnesota Population Center’s Integrated Public Use Microdata Series (IPUMS).
For general demographic information, we use data from the 2017 Annual Social and Economic Supplement (ASEC) for the CPS. From 2002–2015, the CPS included a Volunteer Supplement every September, so we use 2015 data for data on volunteering and donating to charity.
IPUMS requires that you manually generate a data extract through their website, so downloading data from them is not entirely automated or reproducible. We created two extracts (though this could have been combined into one), with the following variables
"data/raw_data/ipums-cps/cps_2017.dat.gz": 2017 ASEC, with the following variables selected (in addition to whatever IPUMS preselects by default) (and weighted byASECWT):AGESEXEDUCINCTOT
"data/raw_data/ipums-cps/cps_09_2015.dat.gz": September 2015 basic monthly CPS (which includes the Volunteer Supplement), with the following variables selected (and weighted byVLSUPPWT):VLSTATUSVLDONATE
We do not show other respondent demographic details because we don’t have good population-level data to compare our sample with. We could theoretically use Pew data for political preferences, but they collect data on party affiliation, while we collected data about respondent positions along a conservative–liberal spectrum, so the two variables aren’t comparable.
cps_2015_ddi_file <- here("data", "raw_data", "ipums-cps", "cps_09_2015.xml")
cps_2015_data_file <- here("data", "raw_data", "ipums-cps", "cps_09_2015.dat.gz")
cps_2015_ddi <- read_ipums_ddi(cps_2015_ddi_file)
cps_2015_data <- read_ipums_micro(cps_2015_ddi_file, data_file = cps_2015_data_file, verbose = FALSE)
cps_2017_ddi_file <- here("data", "raw_data", "ipums-cps", "cps_2017.xml")
cps_2017_data_file <- here("data", "raw_data", "ipums-cps", "cps_2017.dat.gz")
cps_2017_ddi <- read_ipums_ddi(cps_2017_ddi_file)
cps_2017_data <- read_ipums_micro(cps_2017_ddi_file, data_file = cps_2017_data_file, verbose = FALSE)
# Volunteering data from September 2015 only
df_volunteering <- cps_2015_data %>%
# Remove values not in the universe
mutate_at(vars(VLSTATUS, VLDONATE), list(~ifelse(. == 99, NA, .)))
# All other data from annual March 2017 survey
df_demographics <- cps_2017_data %>%
# Remove values not in the universe
mutate(SEX = ifelse(SEX == 9, NA, SEX),
EDUC = ifelse(EDUC <= 1 | EDUC == 999, NA, EDUC),
INCTOT = ifelse(INCTOT == 99999999, NA, INCTOT))global_demographics <- df_demographics %>%
summarize(age = weighted.mean(AGE >= 35, ASECWT),
female = weighted.mean(SEX == 2, ASECWT),
college = weighted.mean(EDUC >= 111, ASECWT, na.rm = TRUE),
income = weighted.mean(INCTOT >= 50000, ASECWT, na.rm = TRUE)) %>%
c()
global_vol <- df_volunteering %>%
summarize(volunteering = weighted.mean(VLSTATUS == 2, VLSUPPWT, na.rm = TRUE),
donating = weighted.mean(VLDONATE == 2, VLSUPPWT, na.rm = TRUE)) %>%
c()
global_stats <- c(global_vol, global_demographics)compare_sample_to_pop <- function(sample_value, population_value) {
mcmc_samples <- pop_prop_stan(
x = table(sample_value)[1],
n_total = length(sample_value),
pop_prop = population_value,
chains = CHAINS, iter = ITER, warmup = WARMUP, seed = BAYES_SEED)
tidied <- tidy(mcmc_samples, conf.int = TRUE, conf.level = 0.95,
estimate.method = "median", conf.method = "HPDinterval") %>%
mutate(in_hpdi = (population_value >= conf.low & population_value <= conf.high))
thetas <- unlist(extract(mcmc_samples, "theta"))
pop_quantile_in_sample <- ecdf(thetas)(population_value)
in_hpdi <- (population_value >= tidied[1,]$conf.low &
population_value <= tidied[1,]$conf.high)
return(list(mcmc_samples = mcmc_samples, tidied = tidied, theta_in_hpdi = in_hpdi,
pop_quantile_in_sample = pop_quantile_in_sample))
}
calc_sample_pop <- tribble(
~Variable, ~sample_value, ~National,
"Female (%)^a^", results$gender_bin, global_stats$female,
"Age (% 35+)^a^", results$age_bin, global_stats$age,
"Income (% $50,000+)^a^", results$income_bin, global_stats$income,
"Education (% BA+)^a^", results$education_bin, global_stats$college,
"Donated in past year (%)^b^", results$give_charity_2, global_stats$donating,
"Volunteered in past year (%)^b^", results$volunteer, global_stats$volunteering
) %>%
mutate(Sample = sample_value %>% map_dbl(~ prop.table(table(.))[1]),
prop_test_bayes = map2(.x = sample_value, .y = National,
.f = ~ compare_sample_to_pop(.x, .y))) format_hpdi <- function(post_lower, post_upper, star, digits = 1) {
glue("({lower}%, {upper}%){star}",
lower = round(100 * post_lower, digits),
upper = round(100 * post_upper, digits))
}
tbl_sample_pop <- calc_sample_pop %>%
mutate(in_hpdi = prop_test_bayes %>% map_lgl(~ .$theta_in_hpdi),
not_hpdi_symbol = ifelse(in_hpdi, "", "^†^"),
diffs_tidy = prop_test_bayes %>% map(~ .$tidied[2,]),
diffs_median = diffs_tidy %>% map_dbl(~ .$estimate),
diffs_hpdi_fancy = diffs_tidy %>%
map2_chr(.x = diffs_tidy, .y = not_hpdi_symbol,
.f = ~ format_hpdi(.x$conf.low, .x$conf.high, .y))) %>%
mutate_at(vars(National, Sample, diffs_median), list(percent)) %>%
select(Variable, Sample, National,
`∆~median~` = diffs_median,
`95% HPDI` = diffs_hpdi_fancy)
note_row <- tibble(Variable = c("*^a^Annual CPS, March 2017*",
"*^b^Monthly CPS, September 2015*",
"*^†^National value is outside the sample highest posterior density interval (HPDI)*"))
bind_rows(tbl_sample_pop, note_row) %>%
pandoc.table.return(keep.line.breaks = TRUE, style = "multiline", justify = "lcccc",
caption = "Characteristics of experimental sample {#tbl:exp-sample}") %T>%
cat(file = here("analysis", "output", "tables", "tbl-exp-sample.md")) %>%
cat()| Variable | Sample | National | ∆median | 95% HPDI |
|---|---|---|---|---|
| Female (%)a | 54.80% | 51.0% | 3.7% | (-0.7%, 7.7%) |
| Age (% 35+)a | 47.27% | 53.9% | -6.6% | (-10.7%, -2.2%)† |
| Income (% $50,000+)a | 50.39% | 27.4% | 21.7% | (17.3%, 26%)† |
| Education (% BA+)a | 46.14% | 29.9% | 16.2% | (11.9%, 20.2%)† |
| Donated in past year (%)b | 82.49% | 48.8% | 33.6% | (30.3%, 36.8%)† |
| Volunteered in past year (%)b | 54.24% | 75.1% | -20.9% | (-25%, -16.7%)† |
| aAnnual CPS, March 2017 | ||||
| bMonthly CPS, September 2015 | ||||
| †National value is outside the sample highest posterior density interval (HPDI) |
Results using interaction terms
In our preregistration protocol, we said we’d calculate the differences in group means using a regression model with 3-way interaction terms. This was mostly for the sake of simplicity—running one model is easier than running multiple individual tests—but the interpretation is a complicated mess. So instead, we ran individual difference in mean tests for the actual paper. For the sake of transparency, though, here are the results from the fully interacted regression models.
Interpreting interactions in models
Ordinarily, people use ANOVA to analyze 3-way, 2 × 2 × 2 factorial designs, like this. A more flexible (yet identical!) way to do this is to use a regular regression model with interaction terms for each of the conditions, like so (here we use three, since we can use simper models to get averages for the larger umbrella groups, like just crackdowns and crackdown + issue):
Model 1:
\[ y = \beta_0 + \beta_1 \text{Crackdown} \]
Model 2:
\[ \begin{aligned} y =& \beta_0 + \beta_1 \text{Crackdown} + \beta_2 \text{Issue } + \\ & \beta_3 \text{Crackdown} \times \text{Issue} \end{aligned} \]
Model 3:
\[ \begin{aligned} y =& \beta_0 + \beta_1 \text{Crackdown} + \beta_2 \text{Issue} + \beta_3 \text{Funding } + \\ & \beta_4 \text{Crackdown} \times \text{Issue } + \\ & \beta_5 \text{Crackdown} \times \text{Funding } + \\ & \beta_6 \text{Issue} \times \text{Funding } + \\ & \beta_7 \text{Crackdown} \times \text{Issue} \times \text{Funding} \end{aligned} \]
Adding different combinations of the coefficients provides the average values for each combination of factors, which corresponds to the average likelihood and amount donated.
model1 <- "1"
model2 <- "2"
model3 <- "3"
interpretation <- tribble(
~Model, ~`Coefficients to add together`,
model3, "Intercept",
model3, "Intercept + Funding",
model2, "Intercept",
model3, "Intercept + Issue",
model3, "Intercept + Issue + Funding + (Issue × Funding)",
model2, "Intercept + Issue",
model1, "Intercept",
model3, "Intercept + Crackdown",
model3, "Intercept + Crackdown + Funding + (Crackdown × Funding)",
model2, "Intercept + Crackdown",
model3, "Intercept + Crackdown + Issue + (Crackdown × Issue)",
model3, "Intercept + Crackdown + Issue + Funding + (Crackdown × Issue) + (Crackdown × Funding) + (Issue × Funding) + (Crackdown × Issue × Funding)",
model2, "Intercept + Crackdown + Issue + (Crackdown × Issue)",
model1, "Intercept + Crackdown"
)
conditions_summary <- bind_rows(group_by(results, crackdown, issue, funding) %>% nest(),
group_by(results, crackdown, issue) %>% nest(),
group_by(results, crackdown) %>% nest(),
results %>% nest(data = everything())) %>%
arrange(crackdown, issue, funding) %>%
select(-data) %>% ungroup() %>%
mutate(funding = ifelse(is.na(funding) & !is.na(issue) , "*Total*", as.character(funding)),
issue = ifelse(is.na(issue) & !is.na(crackdown), "*Total*", as.character(issue)),
crackdown = ifelse(is.na(crackdown), "*Total*", as.character(crackdown))) %>%
group_by(crackdown) %>%
mutate(issue = replace(issue, duplicated(issue), NA)) %>%
ungroup() %>%
mutate(crackdown = replace(crackdown, duplicated(crackdown), NA)) %>%
rename(`Crackdown condition` = crackdown, `Issue condition` = issue,
`Funding condition` = funding)
conditions_summary %>%
slice(-n()) %>%
bind_cols(interpretation) %>%
pandoc.table()| Crackdown condition | Issue condition | Funding condition | Model | Coefficients to add together |
|---|---|---|---|---|
| No crackdown | Human rights | Government | 3 | Intercept |
| Private | 3 | Intercept + Funding | ||
| Total | 2 | Intercept | ||
| Humanitarian assistance | Government | 3 | Intercept + Issue | |
| Private | 3 | Intercept + Issue + Funding + (Issue × Funding) | ||
| Total | 2 | Intercept + Issue | ||
| Total | 1 | Intercept | ||
| Crackdown | Human rights | Government | 3 | Intercept + Crackdown |
| Private | 3 | Intercept + Crackdown + Funding + (Crackdown × Funding) | ||
| Total | 2 | Intercept + Crackdown | ||
| Humanitarian assistance | Government | 3 | Intercept + Crackdown + Issue + (Crackdown × Issue) | |
| Private | 3 | Intercept + Crackdown + Issue + Funding + (Crackdown × Issue) + (Crackdown × Funding) + (Issue × Funding) + (Crackdown × Issue × Funding) | ||
| Total | 2 | Intercept + Crackdown + Issue + (Crackdown × Issue) | ||
| Total | 1 | Intercept + Crackdown |
Amount donated
# Basic interaction models
m_amount_c <- stan_glm(amount_donate ~ crackdown,
data = results, family = gaussian(),
prior = cauchy(location = 0, scale = 2.5),
prior_intercept = cauchy(location = 0, scale = 10),
chains = CHAINS, iter = ITER, warmup = WARMUP, seed = BAYES_SEED)
m_amount_ci <- update(m_amount_c, . ~ . + issue + crackdown * issue)
m_amount_cif <- update(m_amount_c, . ~ . +
issue + funding + crackdown * issue + crackdown * funding +
issue * funding + crackdown * issue * funding)
# Models with just issue and just funding
m_amount_i <- update(m_amount_c, . ~ issue)
m_amount_f <- update(m_amount_c, . ~ funding)# Get subset of coefficients
coefs_to_include <- list(m_amount_c, m_amount_ci, m_amount_cif) %>%
map(~ tidy(.)) %>% bind_rows() %>% distinct(term) %>% pull(term)
huxreg(m_amount_c, m_amount_ci, m_amount_cif,
coefs = coefs_to_include,
statistics = c(Observations = "nobs",
`Posterior sample size` = "pss",
Sigma = "sigma"),
stars = NULL) %T>%
print_hux() %>%
to_md(max_width = 100) %>%
cat(file = here("analysis", "output", "tables", "tbl-interactions-amount.md"))| (1) | (2) | (3) | |
|---|---|---|---|
| (Intercept) | 21.045 | 20.978 | 20.698 |
| (1.490) | (1.707) | (1.977) | |
| crackdownCrackdown | 2.502 | 2.053 | 1.801 |
| (2.061) | (2.019) | (2.251) | |
| issueHumanitarian assistance | -0.241 | 0.177 | |
| (1.692) | (1.844) | ||
| crackdownCrackdown:issueHumanitarian assistance | 1.126 | 2.717 | |
| (2.209) | (3.231) | ||
| fundingPrivate | 0.377 | ||
| (1.858) | |||
| crackdownCrackdown:fundingPrivate | 1.078 | ||
| (2.436) | |||
| issueHumanitarian assistance:fundingPrivate | -0.645 | ||
| (2.242) | |||
| crackdownCrackdown:issueHumanitarian assistance:fundingPrivate | -4.638 | ||
| (5.222) | |||
| Observations | 531 | 531 | 531 |
| Posterior sample size | 8000.000 | 8000.000 | 8000.000 |
| Sigma | 25.653 | 25.640 | 25.589 |
Likelihood of donation
# Basic interaction models
# Weakly informative student t priors, since these coefficients are log-odds
# See https://github.com/stan-dev/stan/wiki/Prior-Choice-Recommendations#prior-for-the-regression-coefficients-in-logistic-regression-non-sparse-case and https://arxiv.org/abs/1507.07170
m_likely_c <- stan_glm(donate_likely_bin ~ crackdown,
data = results, family = binomial(link = "logit"),
prior = student_t(3, 0, 2.5),
prior_intercept = student_t(3, 0, 10),
chains = CHAINS, iter = ITER, warmup = WARMUP, seed = BAYES_SEED)
m_likely_ci <- update(m_likely_c, . ~ . + issue + crackdown * issue)
m_likely_cif <- update(m_likely_c, . ~ . +
issue + funding + crackdown * issue + crackdown * funding +
issue * funding + crackdown * issue * funding)
# Models with just issue and just funding
m_likely_i <- update(m_likely_c, . ~ issue)
m_likely_f <- update(m_likely_c, . ~ funding)# Get subset of coefficients
coefs_to_include <- list(m_likely_c, m_likely_ci, m_likely_cif) %>%
map(~ tidy(.)) %>% bind_rows() %>% distinct(term) %>% pull(term)
huxreg(m_likely_c, m_likely_ci, m_likely_cif,
coefs = coefs_to_include,
statistics = c(Observations = "nobs",
`Posterior sample size` = "pss",
Sigma = "sigma"),
stars = NULL) %T>%
print_hux() %>%
to_md(max_width = 100) %>%
cat(file = here("analysis", "output", "tables", "tbl-interactions-likelihood.md"))| (1) | (2) | (3) | |
|---|---|---|---|
| (Intercept) | -0.298 | -0.275 | -0.175 |
| (0.123) | (0.173) | (0.235) | |
| crackdownCrackdown | 0.161 | 0.028 | -0.666 |
| (0.169) | (0.247) | (0.346) | |
| issueHumanitarian assistance | -0.044 | -0.040 | |
| (0.242) | (0.319) | ||
| crackdownCrackdown:issueHumanitarian assistance | 0.261 | 0.910 | |
| (0.336) | (0.483) | ||
| fundingPrivate | -0.245 | ||
| (0.336) | |||
| crackdownCrackdown:fundingPrivate | 1.389 | ||
| (0.471) | |||
| issueHumanitarian assistance:fundingPrivate | 0.025 | ||
| (0.473) | |||
| crackdownCrackdown:issueHumanitarian assistance:fundingPrivate | -1.300 | ||
| (0.657) | |||
| Observations | 531 | 531 | 531 |
| Posterior sample size | 8000.000 | 8000.000 | 8000.000 |
| Sigma | 1.000 | 1.000 | 1.000 |
Collapsing likelihood variable
To simplify our analysis, we collapse our likelihood scale from a 1-5 Likert scale to a binary variable:
collapsed_likelihood <- tribble(
~`Original answer`, ~`Collapsed answer`,
"Extremely likely", "Likely",
"Somewhat likely", "Likely",
"Neither likely nor unlikely", "Unlikely",
"Somewhat unlikely", "Unlikely",
"Extremely unlikely", "Unlikely"
)
collapsed_likelihood %>%
pandoc.table.return(justify = "ll") %T>%
cat(file = here("analysis", "output", "tables", "tbl-collapsed-likelihood.md")) %>%
cat()| Original answer | Collapsed answer |
|---|---|
| Extremely likely | Likely |
| Somewhat likely | Likely |
| Neither likely nor unlikely | Unlikely |
| Somewhat unlikely | Unlikely |
| Extremely unlikely | Unlikely |
To check that the results are internally consistent when collapsed, we ran an ordered probit model (using Stan) to see if the cutpoints follow the distribution of answers, and they do. “Somewhat likely” and above has a cutpoint of > 0.18, meaning that the likelihood is positive on average for both “Somewhat likely” and “Extremely likely.”
collapsed_oprobit <- stan_polr(donate_likely ~ crackdown, data = results,
prior = R2(0.25, what = "mean"), prior_counts = dirichlet(1),
method = "probit",
chains = CHAINS, iter = ITER, warmup = WARMUP, seed = BAYES_SEED)huxreg(collapsed_oprobit,
coefs = c("Crackdown" = "crackdownCrackdown",
"Cutpoint: Extremely unlikely|Somewhat unlikely" =
"Extremely unlikely|Somewhat unlikely",
"Cutpoint: Somewhat unlikely|Neither likely nor unlikely" =
"Somewhat unlikely|Neither likely nor unlikely",
"Cutpoint: Neither likely nor unlikely|Somewhat likely" =
"Neither likely nor unlikely|Somewhat likely",
"Cutpoint: Somewhat likely|Extremely likely" =
"Somewhat likely|Extremely likely"),
statistics = c(Observations = "nobs",
`Posterior sample size` = "pss"),
stars = NULL) %>%
set_align(everywhere, 1, "left") %>%
set_align(everywhere, 2, "center") %>%
set_position("left") %>%
set_caption("Ordered probit regression with donation likelihood as outcome variable {#tbl:probit-likelihood}") %T>%
print_hux() %>%
to_md(max_width = 140) %>%
cat(file = here("analysis", "output", "tables", "tbl-probit-likelihood.md"))| (1) | |
|---|---|
| Crackdown | 0.089 |
| (0.091) | |
| Cutpoint: Extremely unlikely|Somewhat unlikely | -1.314 |
| (0.091) | |
| Cutpoint: Somewhat unlikely|Neither likely nor unlikely | -0.496 |
| (0.075) | |
| Cutpoint: Neither likely nor unlikely|Somewhat likely | 0.181 |
| (0.072) | |
| Cutpoint: Somewhat likely|Extremely likely | 1.416 |
| (0.093) | |
| Observations | 531 |
| Posterior sample size | 8000.000 |
Miscellaneous survey details
Average time to complete survey
time_summary <- results %>%
summarize_at(vars(duration), list(Minimum = min, Maximum = max, Mean = mean,
`Standard deviation` = sd, Median = median)) %>%
pivot_longer(everything(), names_to = "Statistic", values_to = "value") %>%
mutate(Minutes = fmt_seconds(value)) %>%
select(-value)
pandoc.table(time_summary)| Statistic | Minutes |
|---|---|
| Minimum | 00:49 |
| Maximum | 17:34 |
| Mean | 03:23 |
| Standard deviation | 02:05 |
| Median | 02:48 |
summary_stats <- tableGrob(time_summary, rows = NULL, theme = theme_ngos_table) %>%
gtable::gtable_add_grob(., grobs = rectGrob(gp = gpar(fill = NA, lwd = 1)),
t = 1, b = nrow(.), l = 1, r = ncol(.))
plot_avg_time <- ggplot(results, aes(x = duration)) +
geom_histogram(bins = 50, fill = ngo_cols("blue")) +
scale_x_time(labels = fmt_seconds) +
annotation_custom(summary_stats, xmin = 700, xmax = 900, ymin = 30, ymax = 60) +
labs(x = "Minutes spent on experiment", y = "Count") +
theme_ngos(base_size = 9.5) +
theme(panel.grid.minor = element_blank())
plot_avg_time %T>%
print() %T>%
ggsave(., filename = here("analysis", "output", "figures", "avg-time.pdf"),
width = 4, height = 2.25, units = "in", device = cairo_pdf) %>%
ggsave(., filename = here("analysis", "output", "figures", "avg-time.png"),
width = 4, height = 2.25, units = "in", type = "cairo", dpi = 300)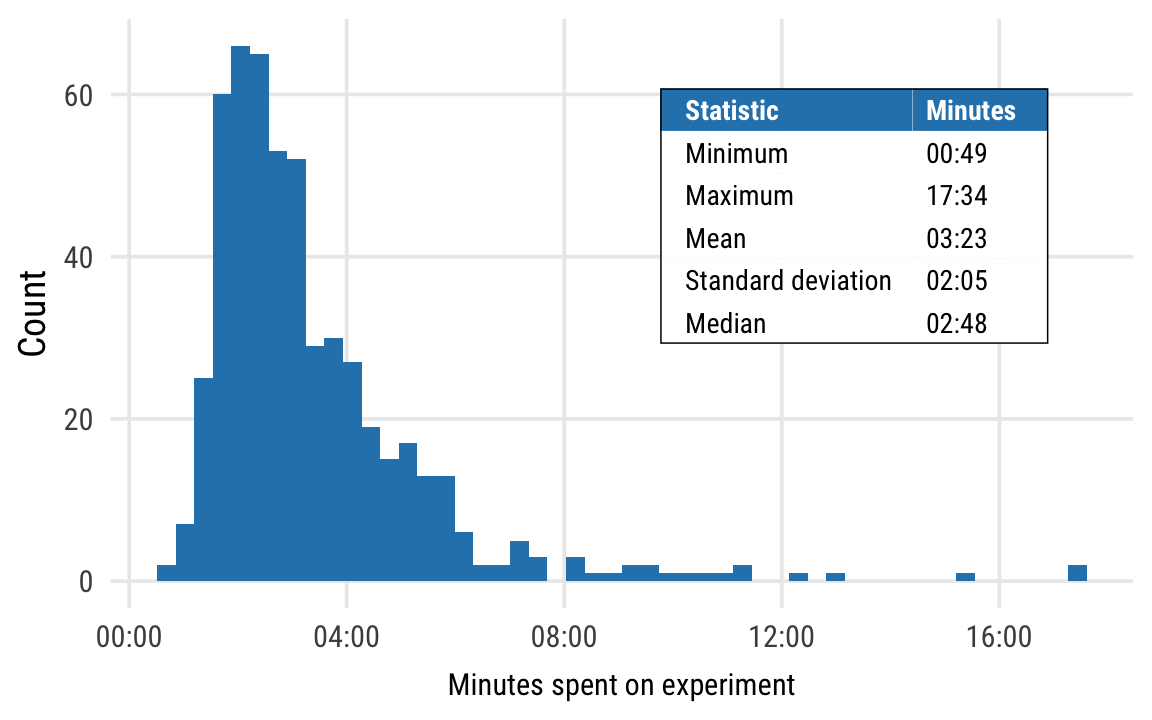
Original computing environment
## # http://dirk.eddelbuettel.com/blog/2017/11/27/#011_faster_package_installation_one
## VER=
## CCACHE=ccache
## CC=$(CCACHE) gcc$(VER)
## CXX=$(CCACHE) g++$(VER)
## CXX11=$(CCACHE) g++$(VER)
## CXX14=$(CCACHE) g++$(VER)
## FC=$(CCACHE) gfortran$(VER)
## F77=$(CCACHE) gfortran$(VER)
##
## CXX14FLAGS=-O3 -march=native -mtune=native -fPIC## ─ Session info ───────────────────────────────────────────────────────────────
## setting value
## version R version 4.0.2 (2020-06-22)
## os macOS Catalina 10.15.6
## system x86_64, darwin17.0
## ui X11
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz America/New_York
## date 2020-10-01
##
## ─ Packages ───────────────────────────────────────────────────────────────────
## package * version date lib
## abind 1.4-5 2016-07-21 [1]
## assertthat 0.2.1 2019-03-21 [1]
## backports 1.1.9 2020-08-24 [1]
## base64enc 0.1-3 2015-07-28 [1]
## bayesplot 1.7.2 2020-05-28 [1]
## blob 1.2.1 2020-01-20 [1]
## boot 1.3-25 2020-04-26 [1]
## broom * 0.7.0 2020-07-09 [1]
## broom.mixed * 0.2.7 2020-09-05 [1]
## callr 3.4.3 2020-03-28 [1]
## cellranger 1.1.0 2016-07-27 [1]
## checkmate 2.0.0 2020-02-06 [1]
## cli 2.0.2 2020-02-28 [1]
## cluster 2.1.0 2019-06-19 [1]
## coda 0.19-3 2019-07-05 [1]
## codetools 0.2-16 2018-12-24 [1]
## colorspace 1.4-1 2019-03-18 [1]
## colourpicker 1.0 2017-09-27 [1]
## commonmark 1.7 2018-12-01 [1]
## crackdownsphilanthropy * 0.9 2020-10-01 [1]
## crayon 1.3.4 2017-09-16 [1]
## crosstalk 1.1.0.1 2020-03-13 [1]
## curl 4.3 2019-12-02 [1]
## data.table 1.13.0 2020-07-24 [1]
## DBI 1.1.0 2019-12-15 [1]
## dbplyr 1.4.4 2020-05-27 [1]
## desc 1.2.0 2018-05-01 [1]
## devtools 2.3.1 2020-07-21 [1]
## digest 0.6.25 2020-02-23 [1]
## dplyr * 1.0.2 2020-08-18 [1]
## DT 0.15 2020-08-05 [1]
## dygraphs 1.1.1.6 2018-07-11 [1]
## ellipsis 0.3.1 2020-05-15 [1]
## evaluate 0.14 2019-05-28 [1]
## fansi 0.4.1 2020-01-08 [1]
## farver 2.0.3 2020-01-16 [1]
## fastmap 1.0.1 2019-10-08 [1]
## forcats * 0.5.0 2020-03-01 [1]
## foreign 0.8-80 2020-05-24 [1]
## forestplot 1.10 2020-07-16 [1]
## Formula 1.2-3 2018-05-03 [1]
## fs 1.5.0 2020-07-31 [1]
## generics 0.0.2 2018-11-29 [1]
## ggplot2 * 3.3.2 2020-06-19 [1]
## ggridges 0.5.2 2020-01-12 [1]
## glue * 1.4.2 2020-08-27 [1]
## Gmisc * 1.11.0 2020-07-03 [1]
## gridExtra * 2.3 2017-09-09 [1]
## gtable 0.3.0 2019-03-25 [1]
## gtools 3.8.2 2020-03-31 [1]
## haven 2.3.1 2020-06-01 [1]
## here * 0.1 2017-05-28 [1]
## hipread 0.2.2 2020-04-29 [1]
## Hmisc 4.4-1 2020-08-10 [1]
## hms 0.5.3 2020-01-08 [1]
## htmlTable * 2.0.1 2020-07-05 [1]
## htmltools 0.5.0 2020-06-16 [1]
## htmlwidgets 1.5.1 2019-10-08 [1]
## httpuv 1.5.4 2020-06-06 [1]
## httr 1.4.2 2020-07-20 [1]
## huxtable * 5.0.0 2020-06-15 [1]
## igraph 1.2.5 2020-03-19 [1]
## inline 0.3.15 2018-05-18 [1]
## ipumsr * 0.4.5 2020-07-21 [1]
## jpeg 0.1-8.1 2019-10-24 [1]
## jsonlite 1.7.0 2020-06-25 [1]
## knitr 1.29 2020-06-23 [1]
## labeling 0.3 2014-08-23 [1]
## later 1.1.0.1 2020-06-05 [1]
## lattice 0.20-41 2020-04-02 [1]
## latticeExtra 0.6-29 2019-12-19 [1]
## lifecycle 0.2.0 2020-03-06 [1]
## lme4 1.1-23 2020-04-07 [1]
## loo 2.3.1 2020-07-14 [1]
## lubridate 1.7.9 2020-06-08 [1]
## magrittr * 1.5 2014-11-22 [1]
## markdown 1.1 2019-08-07 [1]
## MASS 7.3-52 2020-08-18 [1]
## Matrix 1.2-18 2019-11-27 [1]
## matrixStats 0.56.0 2020-03-13 [1]
## memoise 1.1.0 2017-04-21 [1]
## mime 0.9 2020-02-04 [1]
## miniUI 0.1.1.1 2018-05-18 [1]
## minqa 1.2.4 2014-10-09 [1]
## modelr 0.1.8 2020-05-19 [1]
## munsell 0.5.0 2018-06-12 [1]
## nlme 3.1-149 2020-08-23 [1]
## nloptr 1.2.2.2 2020-07-02 [1]
## nnet 7.3-14 2020-04-26 [1]
## pander * 0.6.3 2018-11-06 [1]
## pillar 1.4.6 2020-07-10 [1]
## pkgbuild 1.1.0 2020-07-13 [1]
## pkgconfig 2.0.3 2019-09-22 [1]
## pkgload 1.1.0 2020-05-29 [1]
## plyr 1.8.6 2020-03-03 [1]
## png 0.1-7 2013-12-03 [1]
## prettyunits 1.1.1 2020-01-24 [1]
## processx 3.4.3 2020-07-05 [1]
## promises 1.1.1 2020-06-09 [1]
## ps 1.3.4 2020-08-11 [1]
## purrr * 0.3.4 2020-04-17 [1]
## R6 2.4.1 2019-11-12 [1]
## RColorBrewer 1.1-2 2014-12-07 [1]
## Rcpp * 1.0.5 2020-07-06 [1]
## RcppParallel 5.0.2 2020-06-24 [1]
## readr * 1.3.1 2018-12-21 [1]
## readxl 1.3.1 2019-03-13 [1]
## remotes 2.2.0 2020-07-21 [1]
## reprex 0.3.0 2019-05-16 [1]
## reshape2 1.4.4 2020-04-09 [1]
## rlang 0.4.7 2020-07-09 [1]
## rmarkdown 2.3 2020-06-18 [1]
## rpart 4.1-15 2019-04-12 [1]
## rprojroot 1.3-2 2018-01-03 [1]
## rsconnect 0.8.16 2019-12-13 [1]
## rstan * 2.21.2 2020-07-27 [1]
## rstanarm * 2.21.1 2020-07-20 [1]
## rstantools 2.1.1 2020-07-06 [1]
## rstudioapi 0.11 2020-02-07 [1]
## rvest 0.3.6 2020-07-25 [1]
## scales * 1.1.1 2020-05-11 [1]
## sessioninfo 1.1.1 2018-11-05 [1]
## shiny 1.5.0 2020-06-23 [1]
## shinyjs 1.1 2020-01-13 [1]
## shinystan 2.5.0 2018-05-01 [1]
## shinythemes 1.1.2 2018-11-06 [1]
## StanHeaders * 2.21.0-6 2020-08-16 [1]
## statmod 1.4.34 2020-02-17 [1]
## stringi 1.4.6 2020-02-17 [1]
## stringr * 1.4.0 2019-02-10 [1]
## survival 3.2-3 2020-06-13 [1]
## testthat 2.3.2 2020-03-02 [1]
## threejs 0.3.3 2020-01-21 [1]
## tibble * 3.0.3 2020-07-10 [1]
## tidyr * 1.1.2 2020-08-27 [1]
## tidyselect 1.1.0 2020-05-11 [1]
## tidyverse * 1.3.0 2019-11-21 [1]
## TMB 1.7.18 2020-07-27 [1]
## usethis 1.6.1 2020-04-29 [1]
## V8 3.2.0 2020-06-19 [1]
## vctrs 0.3.4 2020-08-29 [1]
## withr 2.2.0 2020-04-20 [1]
## xfun 0.16 2020-07-24 [1]
## XML 3.99-0.5 2020-07-23 [1]
## xml2 1.3.2 2020-04-23 [1]
## xtable 1.8-4 2019-04-21 [1]
## xts 0.12-0 2020-01-19 [1]
## yaml 2.2.1 2020-02-01 [1]
## zeallot 0.1.0 2018-01-28 [1]
## zoo 1.8-8 2020-05-02 [1]
## source
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.2)
## Github (bbolker/broom.mixed@3d12088)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## local
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.2)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.2)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.2)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.2)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.0)
## CRAN (R 4.0.2)
## CRAN (R 4.0.0)
##
## [1] /Library/Frameworks/R.framework/Versions/4.0/Resources/library