Figures for “Global Assessment Power in the Twenty-First Century”
Judith Kelley and Beth Simmons
July 31, 2018
Data and code
Replication data and code available at GitHub.
library(tidyverse)
library(forcats)
library(stringr)
library(ggstance)
library(ggforce)
library(maptools)
library(rgdal)
library(scales)
library(zoo)
# Useful functions
theme_gpa <- function(base_size=10, base_family="Clear Sans") {
update_geom_defaults("bar", list(fill = "grey30"))
update_geom_defaults("line", list(colour = "grey30"))
update_geom_defaults("label", list(family="Clear Sans"))
update_geom_defaults("text", list(family="Clear Sans"))
ret <- theme_bw(base_size, base_family) +
theme(panel.background = element_rect(fill="#ffffff", colour=NA),
axis.title.y = element_text(margin = margin(r = 10)),
axis.title.x = element_text(margin = margin(t = 10)),
axis.text = element_text(colour="black"),
title=element_text(vjust=1.2, family="Clear Sans", face="bold"),
plot.subtitle=element_text(family="Clear Sans"),
plot.caption=element_text(family="Clear Sans",
size=rel(0.8), colour="grey70"),
panel.border = element_blank(),
axis.line=element_line(colour="grey50", size=0.2),
#panel.grid=element_blank(),
axis.ticks=element_blank(),
legend.position="bottom",
legend.title=element_text(size=rel(0.8)),
axis.title=element_text(size=rel(0.8), family="Clear Sans", face="bold"),
strip.text=element_text(size=rel(1), family="Clear Sans", face="bold"),
strip.background=element_rect(fill="#ffffff", colour=NA),
panel.spacing.y=unit(1.5, "lines"),
legend.key=element_blank(),
legend.spacing=unit(0.2, "lines"))
ret
}
theme_blank_map <- function(base_size=9.5, base_family="Clear Sans") {
ret <- theme_bw(base_size, base_family) +
theme(panel.background = element_rect(fill="#ffffff", colour=NA),
panel.border=element_blank(), axis.line=element_blank(),
panel.grid=element_blank(), axis.ticks=element_blank(),
axis.title=element_blank(), axis.text=element_blank())
ret
}
fig.save.cairo <- function(fig, filepath=file.path(PROJHOME, "Output"),
filename, width, height, units="in", seed=NULL, ...) {
if (!is.null(seed)) set.seed(seed)
ggsave(fig, filename=file.path(filepath, paste0(filename, ".pdf")),
width=width, height=height, units=units, device=cairo_pdf, ...)
if (!is.null(seed)) set.seed(seed)
ggsave(fig, filename=file.path(filepath, paste0(filename, ".png")),
width=width, height=height, units=units, type="cairo", dpi=300, ...)
}
# Clean up the interval labels created by cut()
# Use with forcats::fct_relabel
clean.cut.range <- function(x) {
# If the level starts with "(", strip all the ( and [s and add 1 to the first value
need.to.clean <- str_detect(x, "^\\(")
cleaned <- str_replace_all(x[need.to.clean], "\\(|\\]", "") %>%
map_chr(function(x) {
x.split <- as.numeric(str_split(x, ",", simplify=TRUE))
paste0(x.split[1] + 1, "–", x.split[2])
})
}
my_percent <- percent_format(accuracy = 1)
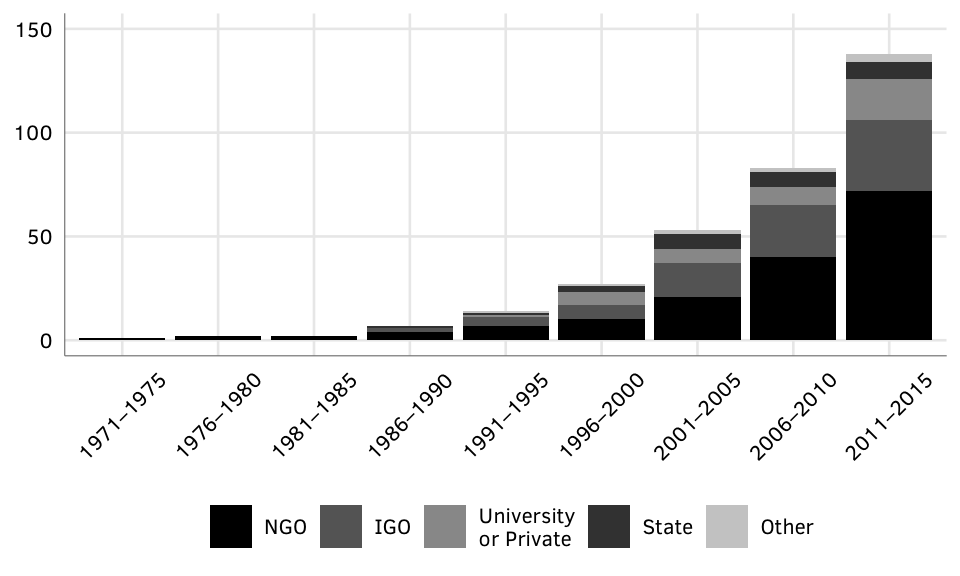
# Load clean dataFigure 1: Cumulative number of GPAs.
year.chunks <- tribble(
~chunk_start, ~chunk_end, ~chunk_name,
1900, 1969, "Pre-1970",
1970, 1974, "1970–74",
1975, 1979, "1975–79",
1980, 1984, "1980–84",
1985, 1989, "1985–89",
1990, 1994, "1990–94",
1995, 1999, "1995–99",
2000, 2004, "2000–04",
2005, 2009, "2005–09",
2010, 2014, "2010–14",
2015, 2020, "2015+"
) %>%
mutate(chunk_name = ordered(fct_inorder(chunk_name)))
year.chunks.long <- year.chunks %>%
rowwise() %>%
summarise(chunk_name = chunk_name, years = list(chunk_start:chunk_end)) %>%
unnest()
years.active <- gpa.data.clean %>%
filter(!is.na(start_year)) %>%
rowwise() %>%
summarise(gpa_id = gpa_id, years = list(start_year:recent_year),
last.active = recent_year) %>%
unnest() %>%
mutate(active = TRUE)
gpas.active.over.time <- years.active %>%
expand(gpa_id, years) %>%
left_join(years.active, by=c("gpa_id", "years")) %>%
group_by(gpa_id) %>%
mutate(last.active = na.locf(last.active, na.rm=FALSE)) %>%
# Get rid of rows with absolutely no data
filter((!is.na(last.active) | !is.na(active))) %>%
# Carry forward the active status of the last year marked if the GPA is still
# active (i.e. last_active is >= 2014). Otherwise, mark as inactive and carry
# forward.
ungroup() %>%
mutate(imputed.active = case_when(
is.na(.$active) & .$last.active >= 2014 ~ TRUE,
is.na(.$active) & .$last.active < 2014 ~ FALSE,
TRUE ~ .$active
)) %>%
# Keep all active years; only keep the first inactive year
group_by(gpa_id, imputed.active) %>%
filter(imputed.active == TRUE |
(imputed.active == FALSE & row_number() == 1)) %>%
ungroup()
gpa.cum.plot <- gpas.active.over.time %>%
left_join(year.chunks.long, by="years") %>%
group_by(gpa_id, chunk_name, imputed.active) %>%
slice(1) %>%
group_by(chunk_name, imputed.active) %>%
summarise(num = n()) %>%
group_by(imputed.active) %>%
# Calculate cumulative total
mutate(cum_total = cumsum(num)) %>%
ungroup() %>%
# Only use cumulative total for inactive ones, since they drop out of the
# data and aren't repeated like the active ones
mutate(plot_value = ifelse(imputed.active, num, cum_total)) %>%
mutate(active = factor(imputed.active, levels=c(TRUE, FALSE),
labels=c("Active GPIs in period", "Cumulative discontinued GPIs"),
ordered=TRUE)) %>%
filter(chunk_name != "2015+")
gpas.active <- gpa.data.clean %>% filter(active == TRUE) %>% nrow
gpas.defunct <- gpa.data.clean %>% filter(active == FALSE) %>% nrowSource: Authors’ database.
Note: “Active” denotes GPAs that were maintained in the given time period. GPAs that have not been updated since 2014 are marked as “Discontinued” in the year following the last active year.
N = 138 active GPAs in 2015; 21 total discontinued GPAs.
fig.cum.gpas <- ggplot(gpa.cum.plot, aes(x=chunk_name, y=plot_value)) +
geom_col(data=filter(gpa.cum.plot, imputed.active), aes(fill=active)) +
geom_line(data=filter(gpa.cum.plot, !imputed.active),
aes(color=active, group=active), size=1) +
scale_color_manual(values=c("grey30"), name=NULL) +
scale_fill_manual(values=c("grey70"), name=NULL) +
guides(fill=guide_legend(order=1),
color=guide_legend(order=2)) +
labs(x=NULL, y=NULL) +
theme_gpa(9) + theme(legend.key.size=unit(0.65, "lines"),
legend.key=element_blank(), legend.spacing=unit(0, "lines"),
legend.margin = margin(0),
panel.grid.major.x=element_blank(),
panel.grid.minor.y = element_blank(),
legend.position = c(0.175, 0.9),
legend.background=element_blank())
fig.cum.gpas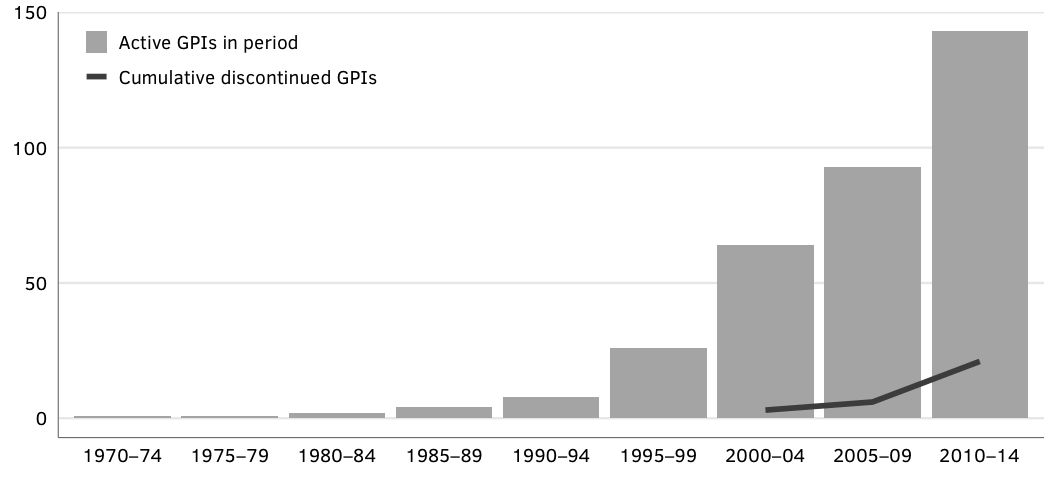
Collapsed subjects
Several of the smaller subject areas are collapsed into larger overarching categories, listed in the table below:
subjects_collapsed %>%
select(`Subject (collapsed)` = subject_collapsed,
`Subject (original)` = subject_area) %>%
pander::pandoc.table(justify = "ll")| Subject (collapsed) | Subject (original) |
|---|---|
| Education | Education |
| Development | Development |
| Economic | Economic |
| Environment | Environment |
| Environment | Energy |
| Governance | Governance |
| Human Rights | Gender |
| Human Rights | Human Rights |
| Human Rights | Press Freedom |
| Security | Security |
| Social | Social |
| Social | Health |
| Trade & Finance | Finance |
| Trade & Finance | Technology |
| Trade & Finance | Trade |
Figure 2 (new): Number of GPAs, by issue and creator type.
gpa.issues.creators <- gpa.data.clean %>%
filter(active == 1, !is.na(subject_collapsed), !is.na(creator_collapsed)) %>%
mutate(subject_collapsed = str_split(subject_collapsed, ", ")) %>%
unnest(subject_collapsed) %>%
group_by(creator_collapsed, subject_collapsed) %>%
summarise(num = n()) %>%
group_by(subject_collapsed) %>%
mutate(total_in_group = sum(num)) %>%
ungroup() %>%
complete(subject_collapsed, creator_collapsed, fill=list(num = 0)) %>%
arrange(total_in_group, num) %>%
mutate(creator_collapsed = fct_relevel(creator_collapsed,
c("Other", "University or Private",
"State", "IGO", "NGO"))) %>%
mutate(subject_collapsed = fct_inorder(subject_collapsed))
issue.creator.denominator <- gpa.data.clean %>%
filter(active == 1, !is.na(creator_collapsed)) %>%
nrowSource: Authors’ database.
Note: Includes only “active” GPAs as of 2014; excludes defunct cases. Note that the total count of GPAs is larger than in Figure 1 because we have double counted cases that straddle issue areas, such as health and development. Overlapping and unknown creators are ommitted.
N = 138.
fig.by.issue.creator <- ggplot(gpa.issues.creators,
aes(x=num, y=subject_collapsed, fill=creator_collapsed)) +
geom_barh(stat="identity", position=position_stackv()) +
scale_x_continuous(sec.axis = sec_axis(~ . / issue.creator.denominator,
labels=my_percent)) +
# scale_fill_grey(start=0.9, end=0.2) +
scale_fill_manual(values=rev(c("black", "grey50", "grey70", "grey25", "grey90"))) +
guides(fill=guide_legend(reverse=TRUE, title=NULL, nrow=1)) +
labs(x=NULL, y=NULL) +
theme_gpa() + theme(panel.grid.major.y=element_blank())
fig.by.issue.creator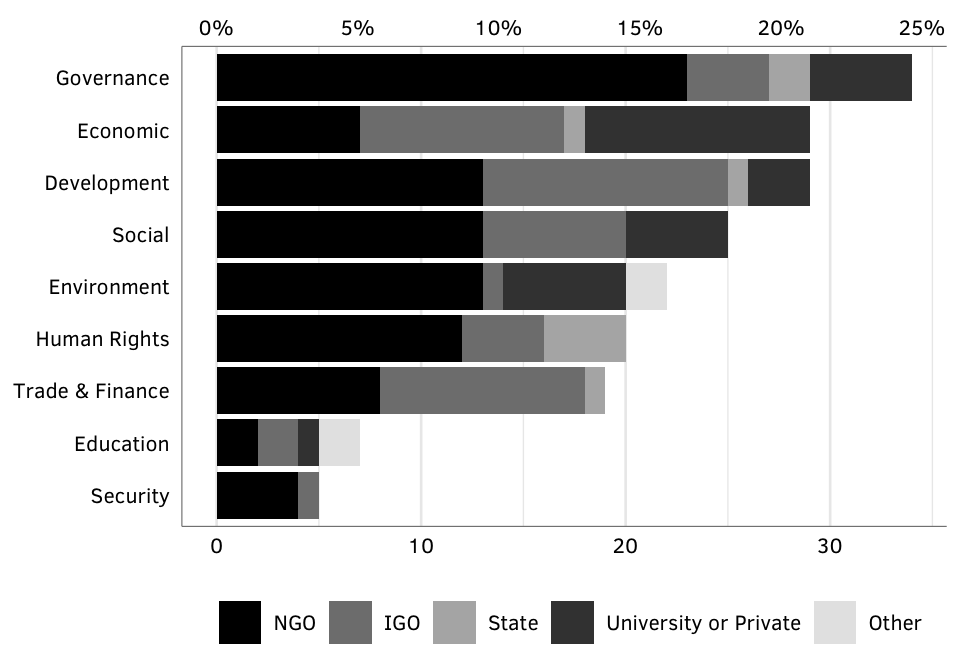
Figure 2: Number of GPAs, by issue.
gpa.issues <- gpa.data.clean %>%
filter(active == 1, !is.na(subject_collapsed)) %>%
mutate(subject_collapsed = str_split(subject_collapsed, ", ")) %>%
unnest(subject_collapsed) %>%
# Get a count of each collapsed issue
group_by(subject_collapsed) %>%
summarise(issue_count = n()) %>%
ungroup() %>%
arrange(issue_count) %>%
mutate(subject_collapsed = fct_inorder(subject_collapsed))
issue.denominator <- gpa.data.clean %>%
filter(active == 1, !is.na(subject_collapsed)) %>%
nrowSource: Authors’ database.
Note: Includes only “active” GPAs as of 2012; excludes defunct cases. Note that the total count of GPAs is larger than in Figure 1 because we have double counted cases that straddle issue areas, such as health and development.
N = 138 active GPAs.
fig.by.issue <- ggplot(gpa.issues, aes(x=issue_count, y=subject_collapsed)) +
geom_barh(stat="identity") +
scale_x_continuous(sec.axis = sec_axis(~ . / issue.denominator,
labels=my_percent)) +
labs(x=NULL, y=NULL) +
theme_gpa() + theme(panel.grid.major.y=element_blank())
fig.by.issue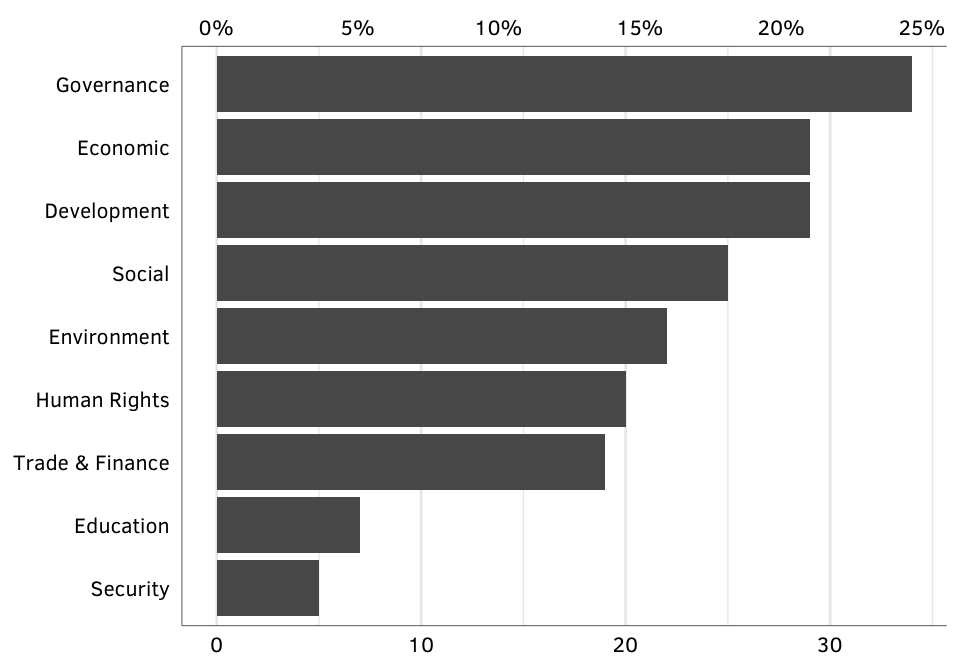
Figure 3: GPA creators, by type.
gpa.creators <- gpa.data.clean %>%
filter(!is.na(creator_collapsed)) %>%
group_by(creator_collapsed) %>%
summarise(num = n()) %>%
arrange(num) %>%
mutate(creator_collapsed = str_replace(creator_collapsed,
"ping or", "ping\nor")) %>%
mutate(creator_collapsed = fct_inorder(creator_collapsed))
creator.denominator <- gpa.data.clean %>%
filter(!is.na(creator_collapsed)) %>%
nrowSource: Authors’ database.
N = 159.
fig.by.creator <- ggplot(gpa.creators, aes(x=num, y=creator_collapsed)) +
geom_barh(stat="identity") +
scale_x_continuous(sec.axis = sec_axis(~ . / creator.denominator,
labels=my_percent)) +
labs(x=NULL, y=NULL) +
theme_gpa() + theme(panel.grid.major.y=element_blank())
fig.by.creator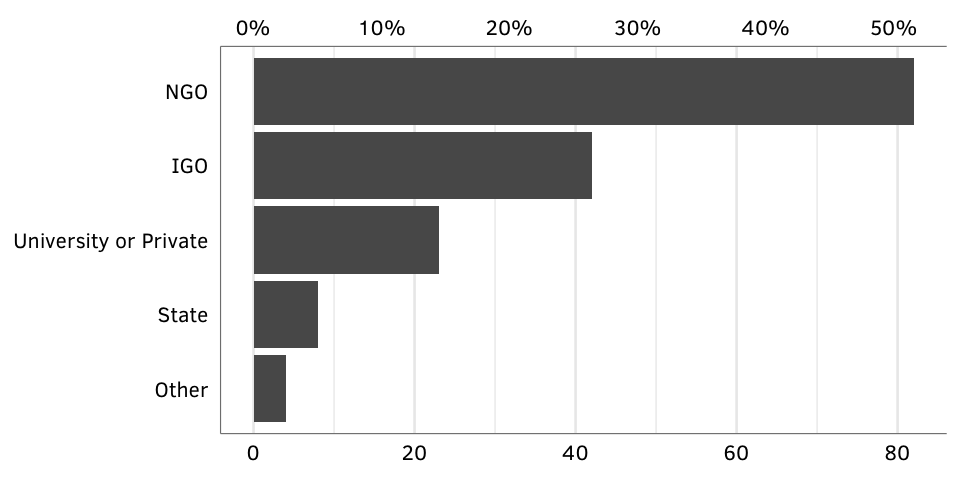
Figure X: GPA creators, by type (GPAs active as of 2014)
gpa.creators.active <- gpa.data.clean %>%
filter(active) %>%
filter(!is.na(creator_collapsed)) %>%
group_by(creator_collapsed) %>%
summarise(num = n()) %>%
arrange(num) %>%
mutate(creator_collapsed = str_replace(creator_collapsed,
"ping or", "ping\nor")) %>%
mutate(creator_collapsed = fct_inorder(creator_collapsed))
creator.active.denominator <- gpa.data.clean %>%
filter(active) %>%
filter(!is.na(creator_collapsed)) %>%
nrow
creator.ngos <- gpa.creators.active %>%
filter(creator_collapsed == "NGO") %>%
select(num) %>% unname() %>% unlist() %>% c()
creator.ngos.pct <- my_percent(creator.ngos /
creator.active.denominator)
creator.states.igos <- gpa.creators.active %>%
filter(creator_collapsed %in% c("IGO", "State")) %>%
summarise(num = sum(num)) %>%
select(num) %>% unname() %>% unlist() %>% c()
creator.states.igos.pct <- my_percent(creator.states.igos /
creator.active.denominator)The figure below shows the distribution of the creator types among the currently active GPAs in our database, showing that 52% percent of GPAs are created by NGOs, while only 30% are in the direct control of states or IGOs.
Source: Authors’ database.
N = 138.
fig.by.creator.active <- ggplot(gpa.creators.active, aes(x=num, y=creator_collapsed)) +
geom_barh(stat="identity") +
scale_x_continuous(sec.axis = sec_axis(~ . / creator.active.denominator,
labels=my_percent)) +
labs(x=NULL, y=NULL) +
theme_gpa() + theme(panel.grid.major.y=element_blank())
fig.by.creator.active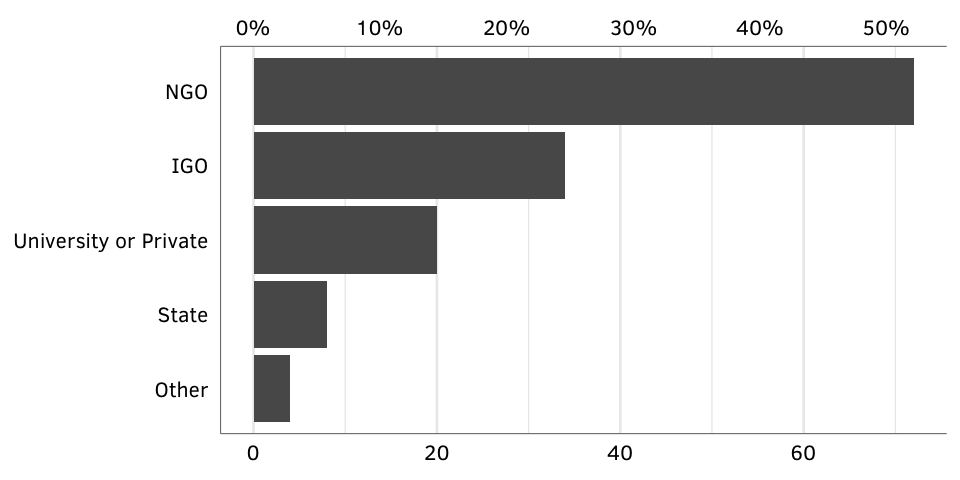
Figure 4: Country of GPA source headquarters.
gpa.countries <- gpa.data.clean %>%
group_by(country_collapsed) %>%
summarise(num = n()) %>%
filter(!is.na(country_collapsed), country_collapsed != "Unknown") %>%
arrange(num) %>%
mutate(country_collapsed = fct_inorder(country_collapsed))
country.denominator <- sum(gpa.countries$num)Source: Authors’ database.
N = 148.
fig.by.country <- ggplot(gpa.countries, aes(x=num, y=country_collapsed)) +
geom_barh(stat="identity") +
scale_x_continuous(sec.axis = sec_axis(~ . / country.denominator,
labels=my_percent)) +
labs(x=NULL, y=NULL) +
theme_gpa() + theme(panel.grid.major.y=element_blank())
fig.by.country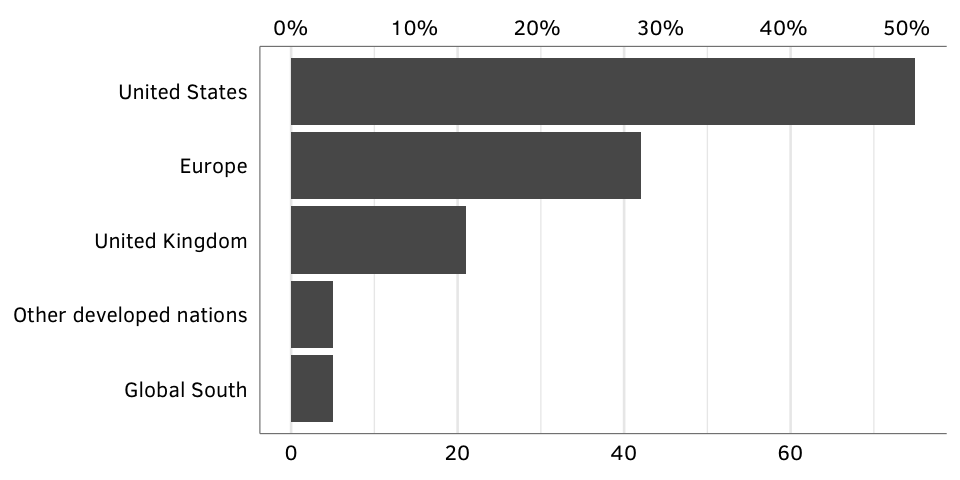
Figure 4: Country of GPA source headquarters (maps).
# Load map data
if (!file.exists(file.path(PROJHOME, "Data", "map_data",
"ne_110m_admin_0_countries.VERSION.txt"))) {
map.url <- paste0("http://www.naturalearthdata.com/",
"http//www.naturalearthdata.com/download/110m/cultural/",
"ne_110m_admin_0_countries.zip")
map.tmp <- file.path(PROJHOME, "Data", basename(map.url))
download.file(map.url, map.tmp)
unzip(map.tmp, exdir=file.path(PROJHOME, "Data", "map_data"))
unlink(map.tmp)
}countries.map <- readOGR(file.path(PROJHOME, "Data", "map_data"),
"ne_110m_admin_0_countries",
verbose=FALSE)
countries.robinson <- spTransform(countries.map, CRS("+proj=robin"))
countries.ggmap <- fortify(countries.robinson, region="iso_a3") %>%
filter(!(id %in% c("ATA", -99))) %>% # Get rid of Antarctica and NAs
mutate(id = ifelse(id == "GRL", "DNK", id)) # Greenland is part of Denmark
possible.countries <- data_frame(ISO3 = unique(as.character(countries.ggmap$id)))
gpa.countries.map <- gpa.data.clean %>%
group_by(ISO3) %>%
summarise(num = n()) %>%
filter(!is.na(ISO3)) %>%
# Bring in full list of countries and create new variable indicating if the
# country has any GPAs
right_join(possible.countries, by="ISO3") %>%
mutate(num.bin = !is.na(num),
num.ceiling = ifelse(num >= 10, 10, num))Source: Authors’ data
N = 17 countries.
gpa.map.bin <- ggplot(gpa.countries.map, aes(fill=num.bin, map_id=ISO3)) +
geom_map(map=countries.ggmap, size=0.15, colour="black") +
expand_limits(x=countries.ggmap$long, y=countries.ggmap$lat) +
coord_equal() +
scale_fill_manual(values = c("white", "grey30"), guide=FALSE) +
theme_blank_map()
gpa.map.bin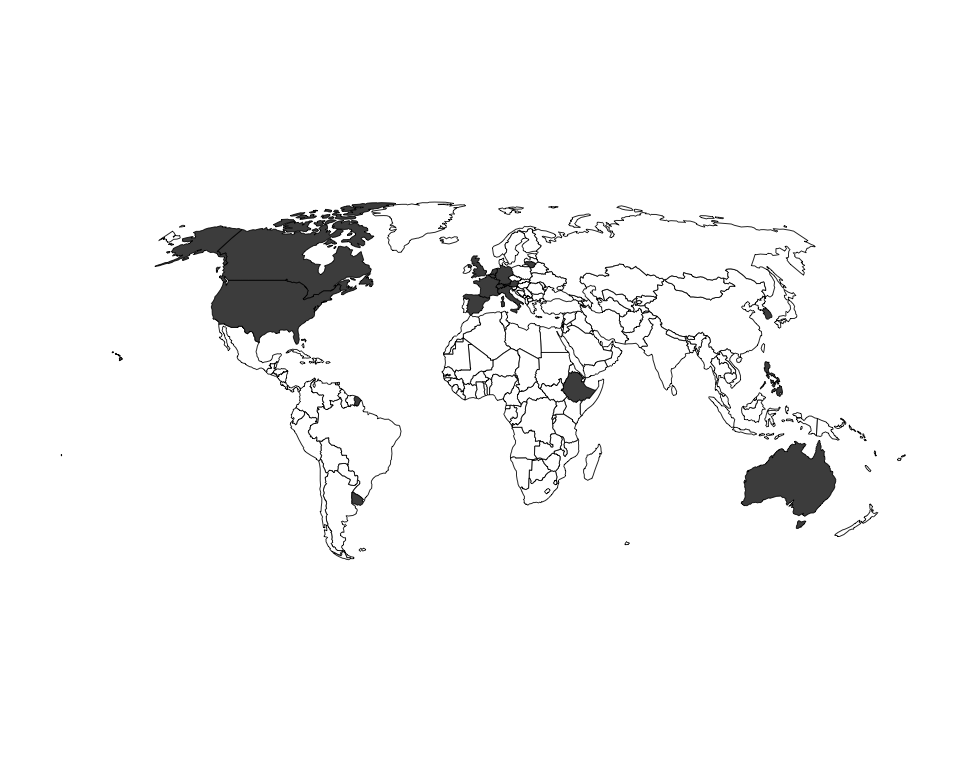
gpa.map <- ggplot(gpa.countries.map, aes(fill=num.ceiling, map_id=ISO3)) +
geom_map(map=countries.ggmap, size=0.15, colour="black") +
expand_limits(x=countries.ggmap$long, y=countries.ggmap$lat) +
coord_equal() +
scale_fill_gradient(low="grey90", high="grey30", breaks=seq(2, 10, 2),
labels=c(paste(seq(2, 8, 2), " "), "10+"),
na.value="white", name="GPAs based in country",
guide=guide_colourbar(ticks=FALSE, barwidth=5)) +
theme_blank_map(9) +
theme(legend.position="bottom", legend.key.size=unit(0.65, "lines"),
strip.background=element_rect(colour="#FFFFFF", fill="#FFFFFF"))
gpa.map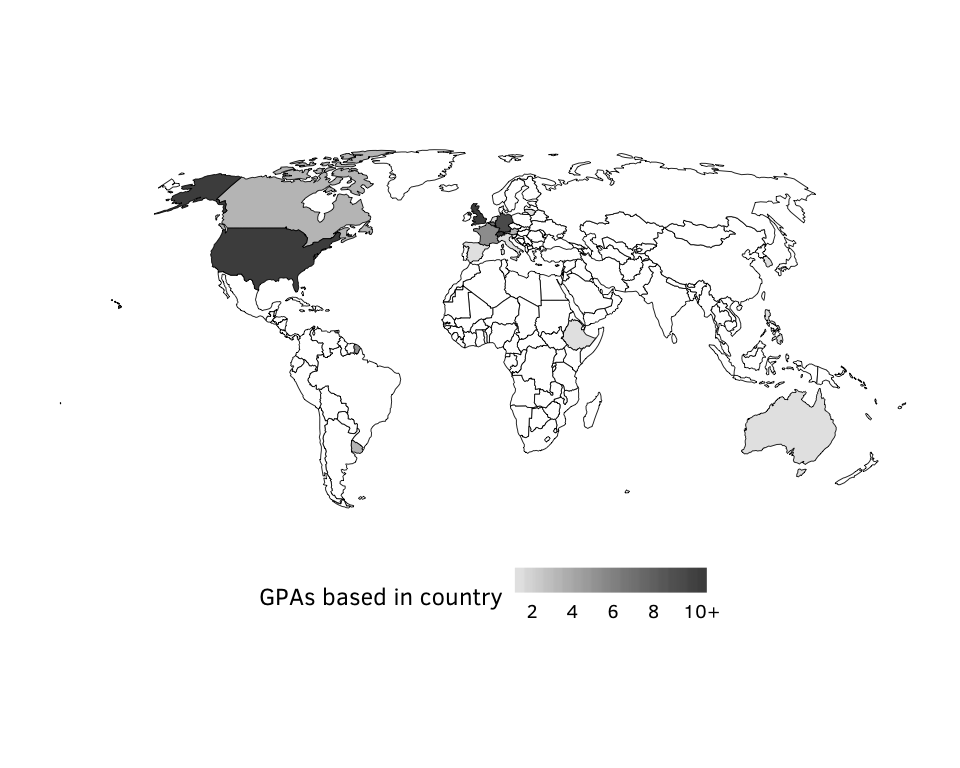
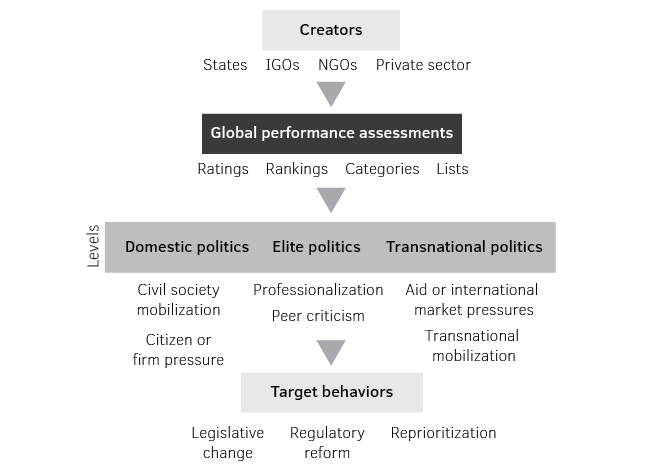
Figure 5: Pathways of GPA influence.
Source: Adapted from Kelley and Simmons, 2015.
(located at ./Output/figure-5-pathways.pdf and ./Output/figure-5-pathways.png)
Figure X: Pseudo-networks of indicator creators over time
Source: Authors’ data
gpa.data.clean.creator.long <- gpa.data.clean %>%
separate_rows(subject_collapsed, sep=",") %>%
mutate(subject_collapsed = str_trim(subject_collapsed))
subject.counts <- gpa.data.clean.creator.long %>%
count(subject_collapsed) %>%
arrange(desc(n))
gpa.creator.cumulative <- gpa.data.clean.creator.long %>%
filter(!is.na(start_year)) %>%
filter(!is.na(creator_collapsed)) %>%
# Infer death year based on most recent year if not active
mutate(end_year = ifelse(active == 0, recent_year, 2015)) %>%
# Create list of years, like 1998:2005
mutate(year = map2(start_year, end_year, ~ seq(.x, .y))) %>%
# Unnest list of years
unnest(year) %>%
filter(year > 1970) %>%
# Divide years into five-year chunks
mutate(pentad = cut(year, breaks=seq(1970, 2015, 5), ordered_result=TRUE),
pentad = fct_relabel(pentad, clean.cut.range)) %>%
# Only select the first year in the pentad. Without this, indexes appear up
# to five times in the pentad.
filter(!is.na(pentad)) %>%
group_by(pentad, gpa_id) %>%
slice(1) %>%
ungroup() %>%
left_join(subject.counts, by="subject_collapsed") %>%
arrange(pentad, desc(n)) %>%
mutate(subject_collapsed = ordered(fct_inorder(subject_collapsed)),
creator_collapsed = ordered(fct_relevel(creator_collapsed,
"NGO", "IGO", "State",
"University or Private",
"Other")))
# Export data
gpa.creator.cumulative %>%
select(gpa_id, gpa_name, start_year, recent_year,
subject = subject_collapsed, creator = creator_collapsed) %>%
arrange(start_year) %>%
write_csv(path = file.path(PROJHOME, "Output", "gpa_creator_cumulative.csv"))
# gpa.creator.cumulative %>%
# count(pentad, subject_collapsed) %>%
# spread(pentad, nn) %>%
# write_csv("~/Desktop/gpa_subject_pentads.csv", na = "")
#
# gpa.creator.cumulative %>%
# count(creator_collapsed, subject_collapsed) %>%
# spread(creator_collapsed, nn) %>%
# write_csv("~/Desktop/gpa_subject_creators.csv", na = "")
#
# gpa.creator.cumulative %>%
# count(pentad, subject_collapsed, creator_collapsed) %>%
# spread(pentad, nn) %>%
# write_csv("~/Desktop/gpa_subject_creator_pentads.csv", na = "")# position_jitternormal() comes from the development version of ggforce, which isn't on CRAN yet.
# Install from GitHub to use: devtools::install_github('thomasp85/ggforce')
gpa.type.points <- ggplot(gpa.creator.cumulative,
aes(x=pentad, y=subject_collapsed,
shape=creator_collapsed)) +
# geom_point(size=0.75, alpha=0.75, position=position_jitter(width=0.25, height=0.30)) +
geom_point(size=0.75, alpha=0.6, position=position_jitternormal(sd_x=0.1, sd_y=0.16)) +
labs(x=NULL, y=NULL) +
guides(shape=guide_legend(title=NULL, override.aes=list(size=3))) +
scale_shape_manual(values=c(16, 21, 17, 24, 3)) +
theme_gpa() +
theme(axis.text.x=element_text(angle=45, hjust=0.5, vjust=0.5),
panel.grid.major=element_blank())
set.seed(12345); gpa.type.points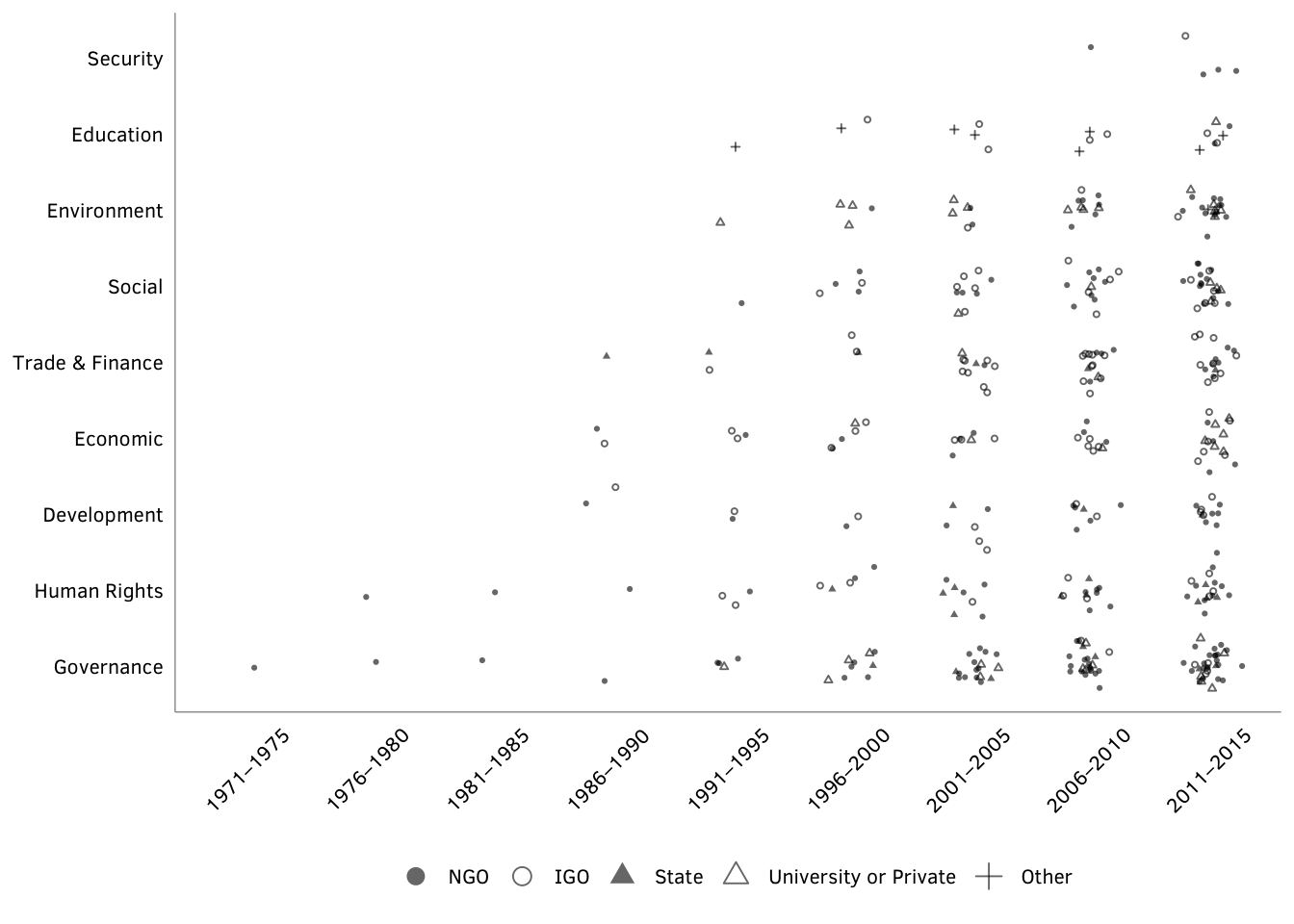
Figure X: Creator types over time
Source: Authors’ data
creator.types.time <- gpa.creator.cumulative %>%
group_by(pentad, creator_collapsed, active) %>%
summarize(n = n()) %>%
filter(active == TRUE) %>%
ungroup() %>%
arrange(desc(n)) %>%
mutate(creator_collapsed = str_replace(creator_collapsed, " or ", "\\\nor ")) %>%
mutate(creator_collapsed = fct_inorder(creator_collapsed))
creator.types.labs <- creator.types.time %>%
group_by(creator_collapsed) %>%
slice(1)
creator.types.lines <- ggplot(creator.types.time,
aes(x = pentad, y = n,
color = creator_collapsed,
group = creator_collapsed)) +
geom_line(size = 1) +
geom_label(data = creator.types.labs, aes(x = pentad, y = n, label = creator_collapsed),
hjust = 0, nudge_x = 0.1, size = 3) +
labs(x=NULL, y=NULL) +
scale_color_manual(values=c("black", "grey25", "grey45", "grey60", "grey70")) +
expand_limits(x = 10.6, y = 80) +
guides(color = FALSE) +
theme_gpa() +
theme(axis.text.x=element_text(angle=45, hjust=0.5, vjust=0.5),
panel.grid.minor=element_blank())
creator.types.lines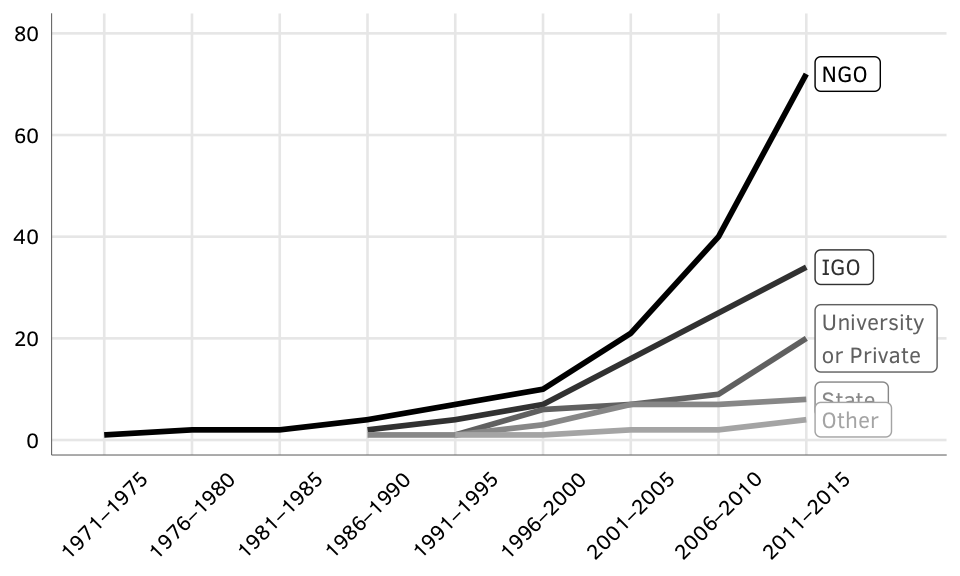
Figure X: Creator types over time
Source: Authors’ data
creator.types.bars <- ggplot(creator.types.time,
aes(x = pentad, y = n, fill = fct_rev(creator_collapsed))) +
geom_bar(stat = "identity") +
labs(x = NULL, y = NULL) +
expand_limits(y = 150) +
scale_fill_manual(values=rev(c("black", "grey40", "grey60", "grey25", "grey80"))) +
guides(fill=guide_legend(reverse=TRUE, title=NULL, nrow=1)) +
theme_gpa() +
theme(axis.text.x=element_text(angle=45, hjust=0.5, vjust=0.5),
panel.grid.minor=element_blank())
creator.types.bars